Figures & data
Table 1. Association between patients’ clinical features and RMRP expression
Figure 1. The expression of RMRP in ESCC. A. RMRP was significantly upregulated in ESCC tissues compared with that in adjacent nontumorous tissues. B. The expression of RMRP was dramatically higher in ESCC cell lines (YES-2, KYSE30, EC 109, and EC9706) than that in the normal esophageal epithelial cell line (HET-1A). ***P < 0.001
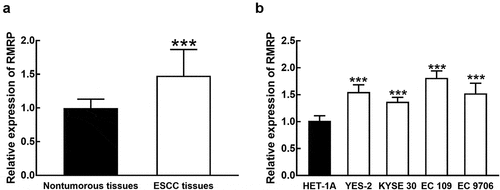
Table 2. Multivariate Cox regression analysis for ESCC patients
Figure 2. Kaplan-Meier curve of ESCC patients with different RMRP expressions. Patients with high RMRP expression possessed poorer survival than patients with low RMRP expression. Log-rank P < 0.001
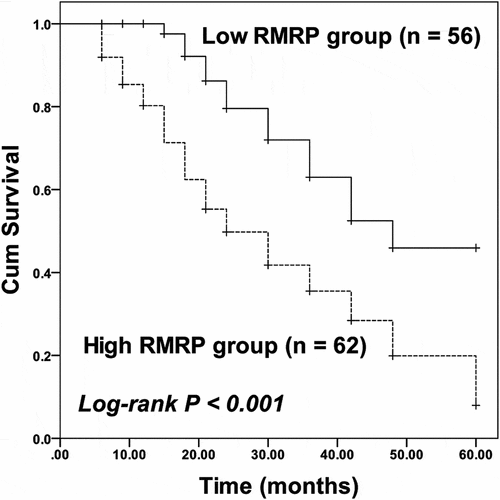
Figure 3. Effect of RMRP expression on cell proliferation of ESCC. A. The expression of RMRP in YES-2 and EC109 cells was significantly inhibited by the transfection of RMRP-siRNA. B. The knockdown of RMRP dramatically suppressed the proliferation of YES-2 and EC109 cells. **P < 0.01, ***P < 0.001
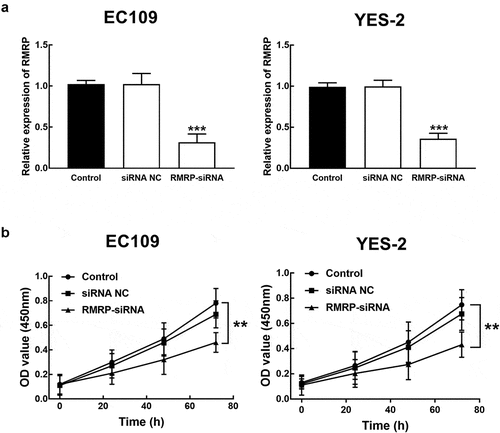
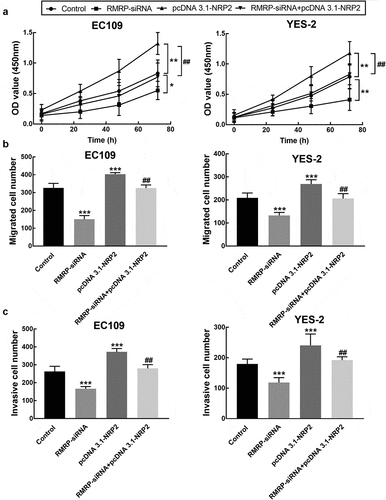
Figure 4. Effect of RMRP expression on cell migration and invasion of ESCC. A. The migration of YES-2 and EC109 cells was significantly inhibited by the knockdown of RMRP. B. The invasion of YES-2 and EC109 cells was significantly inhibited by the silencing of RMRP. ***P < 0.001
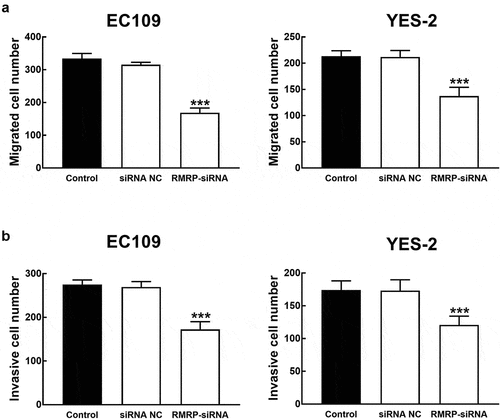
Figure 5. The interaction between RMRP and NRP2. A. The expression of RMRP was positively correlated with NRP2 expression. r = 0.637, P < 0.001. B. The expression level of NRP2 mRNA was significantly suppressed by the knockdown of RMRP and enhanced by the transfection of pcDNA3.1-NRP. The silencing of RMRP could reverse the overexpression of NRP2. C. The expression level of NRP2 protein was dramatically inhibited by the silencing of RMRP and promoted by pcDNA3.1-NRP2. The knockdown could attenuate the upregulation of NRP2. **P < 0.01, ***P < 0.001 relative to the control group. ###P < 0.001 relative to pcDNA3.1-NRP2 group
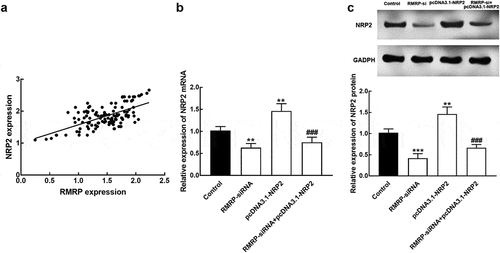
Figure 6. The association of miR-613 with RMRP and NRP2. A. miR-613 overexpression significantly suppressed the luciferase activity of RMR WT, which was enhanced by the knockdown of miR-613. B. miR-613 dramatically regulated the luciferase activity of NRP2 WT, while the NRP2 MT was not affected by miR-613. ***P < 0.001
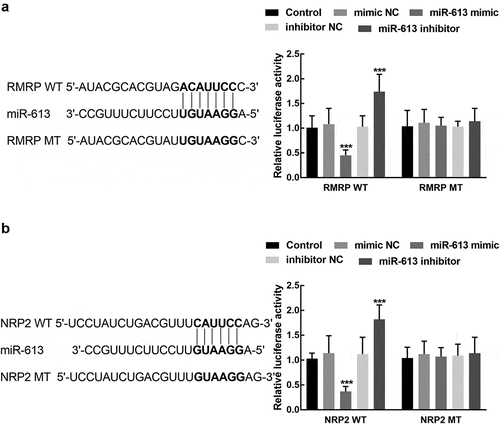
Figure 7. Effect of NRP2 on cell processes of ESCC. A. The overexpression of NRP2 significantly promoted the proliferation of EC109 and YES-2 cells, which was reversed by the knockdown of RMRP. B. The migration of EC109 and YES-2 cells was dramatically improved by NRP2 overexpression, which was reversed by the silencing of RMRP. C. The invasion of EC109 and YES-2 cells was dramatically improved by NRP2 overexpression, which was reversed by the silencing of RMRP. **P < 0.01, ***P < 0.001 relative to the control; ## P < 0.01 relative to pcDNA3.1-NRP2