Figures & data
Figure 1. IGF2-AS expression is down-regulated significantly in HCC cells and tissues, and related with prognosis of HCC patients. IGF2-AS expression was down-regulated in cancer (t) than in normal (n) tissues of HCC patients investigated by bioinformatics assay (a) based on TCGA datasets; (b) analyzed using GEPIA online tool; (c) IGF2-AS expression was dramatically down-regulated in HCC than in adjacent tissues from 106 local HCC patients detected by qRT-PCR assay. (d) IGF2-AS expression was dramatically down-regulated in human HCC (Hep3B, HepG2 and Huh7) than in human normal liver (LO2) cells detected by qRT-PCR, and HepG2 and Huh7 cells had the lowest IGF2-AS expression level. (e) Kaplan-Meier curve showed that low IGF2-AS expression was associated with poor prognosis in overall survival of HCC patients (n = 80). Low IGF2-AS expression was significantly associated with decreased (f) overall survival and (g) disease free survival of HCC patients from the TCGA database. *p < 0.05, **p < 0.01, ***p < 0.01
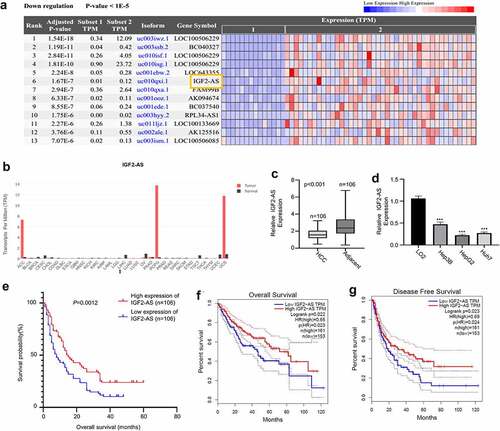
Table 1. Correlations of IGF2-AS expression with clinicopathologic features of hepatocellular carcinoma
Figure 2. IGF2-AS over-expression inhibits proliferation and migration, while promotes apoptosis of HCC cells. IGF2-AS was over-expressed successfully in HepG2 and Huh7 cells (a), then viability (b), colony formation(c), migration(d), invasion(e) and cell apoptosis (f) were investigated and compared between cells with (IGF2-AS) or without (NC) IGF2-AS over-expression. **p < 0.01
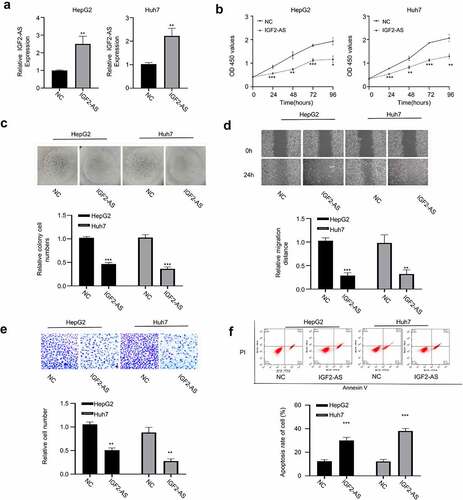
Figure 3. IGF2-AS acts as a sponge for miR-520h. (a) Illustration of prospective miR-520h binding sites in IGF2-AS-3ʹUTR predicted by Starbase 2.0 database and the mutation sites used for specific binding assay (http://starbase.sysu.edu.cn/). (b) miR-520h expression in liver cancer was significantly higher than that of adjacent normal tissues in TCGA database analyzed with the Starbase 2.0 on line tool (http://starbase.sysu.edu.cn/) (p = 0.0017). (c) miR-520h expression in HepG2 and Huh7 cells was significantly higher than that in LO2 cells detected by qRT-PCR. (d) Dual-luciferase reporter assay in HepG2 and Huh7 cells containing wild type (IGF2-AS) or mutated (MUT) reporter of IGF2-AS in the presence of miR-520h mimics or NC. (e) IGF2-AS expression in HepG2 and Huh7 cells after miR-520h was silenced with miR-520h inhibitor or over-expressed with miR-520h mimics. (f) miR-520h expression was dramatically up-regulated in HCC than in adjacent tissues from 106 HCC patients (same specimens as in ). (F) Spearman’s coefficient assay showed a significantly negative association between IGF2-AS and miR-520h expressions in 106 HCC tissues (same specimens as in ). **p < 0.01
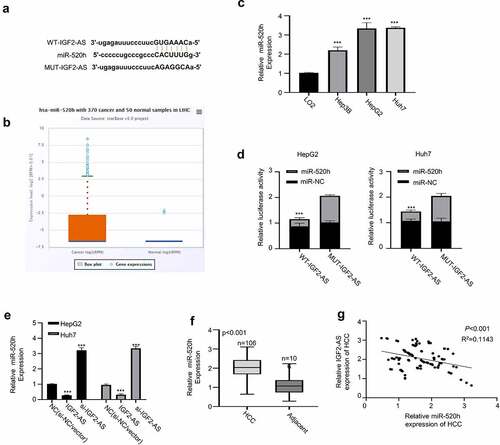
Figure 4. IGF2-AS elevates CDKN1A expression by sponging miR-520h. (a) Diagram of prospective miR-520h binding sites in CDKN1A-3ʹ-UTR predicted with miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php). (b) CDKN1A mRNA expression was significantly down-regulated in HCC (HepG2 and Huh7) than in normal liver (LO2) cells determined by qRT-PCR. (c) dual-luciferase reporter assay in HepG2 and Huh7 cells containing wild type (CDKN1A) or mutated (MUT) reporter of CDKN1A-3ʹ-UTR with miR-520h mimics or negative control (miR-NC). (d) CDKN1A mRNA expression in HepG2 and Huh7 cells after miR-520h was silenced with miR-520h inhibitor or over-expressed with miR-520h mimics. (e) CDKN1A protein expression in HepG2 and Huh7 cells after miR-520h was silenced with miR-520h inhibitor or over-expressed with miR-520h mimics. (f) Spearman’s correlation assay of positive correlations between CDKN1A with IGF2-AS expressions and negative correlations between CDKN1A with miR-520h expressions in 106 HCC tissues (same specimens used in Figure 1C). **p < 0.01
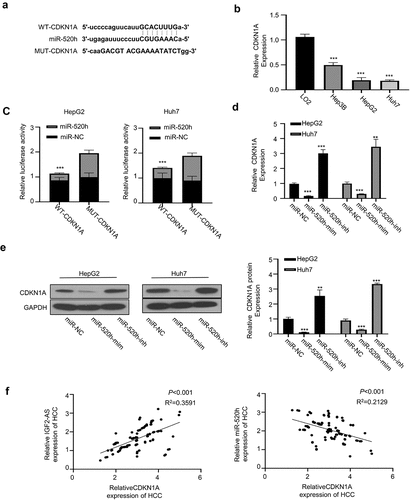
Figure 5. MiR-520h over-expression or CDKN1A silencing reverses IGF2-AS reduced aggressive behaviors of HCC cells. (a) viability, (b) colony formation, (c) migration, and (d) invasion abilities in HepG2 and Huh7 cells after IGF2-AS over-expression (IGF2-AS) with or without miR-520h over-expression by miR-520h mimics (miR-520h) or CDKN1A silencing by si-CDKN1A. **p < 0.01
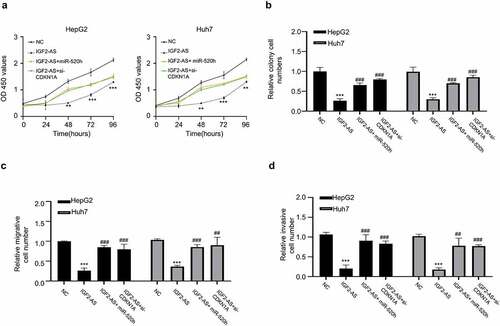
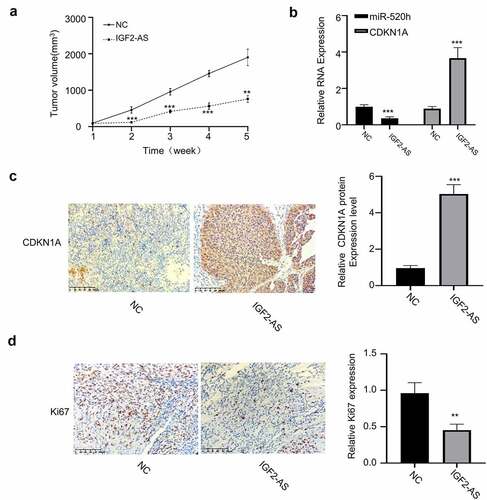
Figure 6. IGF2-AS over-expression in HepG2 cells inhibits xenograft tumor growth by regulating miR-520h/CDKN1A axis in vivo. (a) Tumor volume (n = 5 each). (b) miR-520h and CDKN1A expressions by qRT-PCR assay. (c) CDKN1A and (d) Ki-67 expressions by immunochemistry. **p < 0.01, ***p < 0.001,##p < 0.01, ###p < 0.001