Figures & data
Table 1. Association between SNHG22 or miR-16-5p expression and clinicopathological characteristics in HCC
Figure 1. Relative SNHG22 expression in HCC tissues and cells and its correlation with patients’ survival. (a) RT-qPCR analysis of relative SNHG22 expression in HCC tissues and adjacent non-cancerous tissues. (b) relative SNHG22 expression in HCC cell lines and THLE-3 cell line was detected by RT-qPCR. (c) correlation between SNHG22 expression and HCC patients’ survival. **p < 0.01
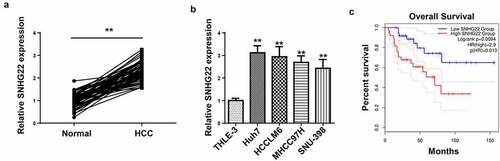
Figure 2. The effect of SNHG22 knockdown on cell proliferation, migration, invasion and angiogenesis. (a) the transfection efficiency of sh-SNHG22 in Huh7 and HCCLM6 cells was examined by RT-qPCR. (b) The proliferative ability of Huh7 and HCCLM6 cells transfected with sh-SNHG22 was determined by CCK-8 assay. (c and d) transwell assay was done for detecting the migration and invasion of transfected Huh7 and HCCLM6 cells. (e) The effect of SNHG22 knockdown on angiogenesis of Huh7 and HCCLM6 cells. (f) western blot showed the protein levels of VEGF and endoglin in HCC cells transfected with sh-NC and sh-SNHG22. **p < 0.01, ***p < 0.001
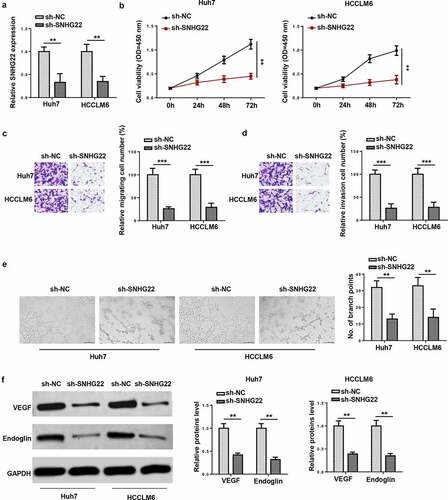
Figure 3. SNHG22 deficiency inhibited tumor growth and angiogenesis. (a) the tumors excised from the nude mice in sh-SNHG22 group or sh-NC group. (b) The volume of tumors in two groups was assessed. (c) the tumor weight from sh-SNHG22 group or sh-NC group. (d) IHC was used for detecting Ki67 expression in each group. (e) MVD was assessed in each group. *p < 0.05, **p < 0.01
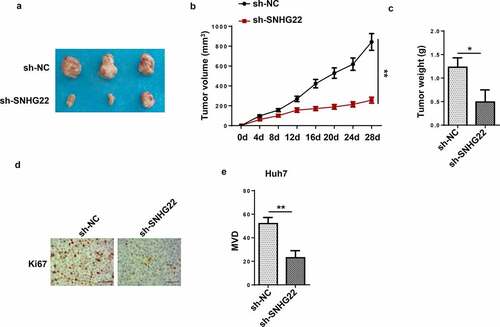
Figure 4. SNHG22 facilitated DNA promoter methylation to inhibit miR-16-5p. (a) The expression of miR-16-5p in HCC tissues and cells. (b) MiR-16-5p expression in HCC cells transfected with sh-SNHG22 or sh-NC. (c) Methylation-specific PCR was conducted to analyze the effect of SNHG22 on miR-16-5p methylation. (d) relative miR-16-5p expression after 5-aza-dC treatment in HCC cells. *p < 0.05, **p < 0.01, ***p < 0.001
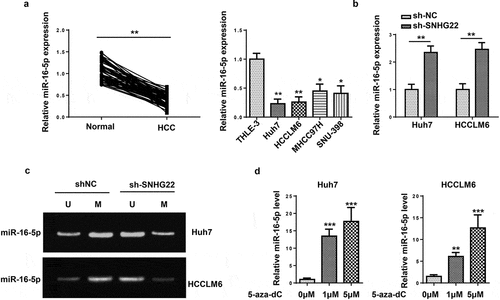
Figure 5. SNHG22 recruited DNMT1 via EZH2 to promote miR-16-5p DNA promoter methylation. (a) the subcellular location of SNHG22 in Huh7 and HCCLM6 cells. (b) RT-qPCR analysis for miR-16-5p expression in cells transfected with sh-DNMT1, sh-DNMT3A and sh-DNMT3B. (c) ChIP-RT-qPCR analysis testified the effect of SNHG22 silencing on the binding of DNMT1 to miR-16-5p promoter. (d) The interaction between SNHG22 and EZH2 was verified by RIP assay. (e) The binding capacity between DNMT1 and EZH2 was validated by Co-IP assay when SNHG22 was silenced. **p < 0.01, ***p < 0.001
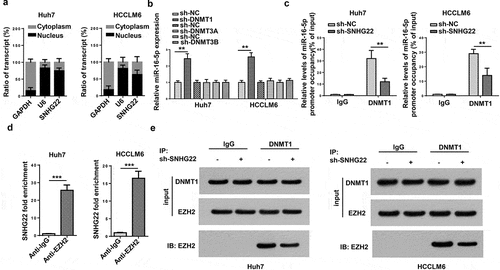
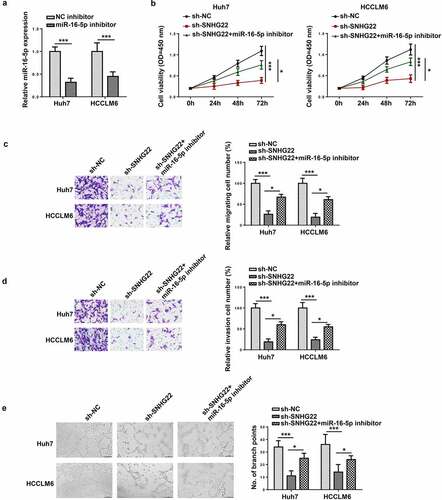
Figure 6. SNHG22 promoted cell proliferation, migration, invasion and angiogenesis via miR-16-5p. (a) RT-qPCR analysis was performed to test the transfection efficiency of miR-16-5p inhibitor. (b) The proliferation of each group was assessed by CCK-8 assay. (c and d) The migration and invasion of the transfected cells were evaluated by transwell assay. (e) Tube formation assay was conducted to verify angiogenesis in each group. *p < 0.05, ***p < 0.001