Figures & data
Figure 1. GNG7 was low expression in LUAD tissue and cell lines (a) mRNA level of GNG7 was detected in 84 pairs of LUAD tissues and normal tissues by RT-qPCR. (b-c) mRNA level of GNG7 in LUAD tissues and normal tissues from TCGA and GEO (GSE19188) database was compared. (d-e) mRNA and protein levels of GNG7 in Lung cancer cell lines (H460, A549, H1299, SK-MES-1 and NCI-H520) and Bronchial epithelial cell line(16HBE) were detected. (f) GNG7 protein expressions in 10 pairs of LUAD. The relative protein levels of GNG7 were normalized against GAPDH. (g) Tissue Microarray with LUAD tissues and normal tissues was used to detected expression of GNG7 by IHC, and the staining score of GNG7 percentage was quantified. * p < 0.05, **p < 0.005
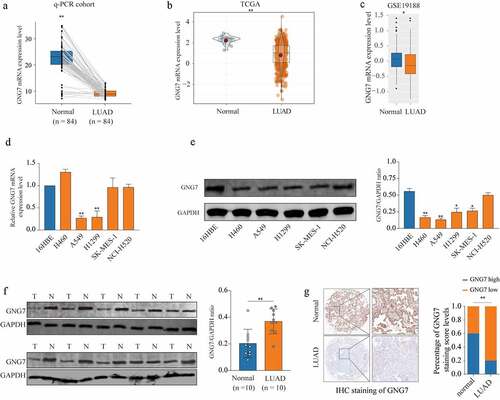
Figure 2. Low expression of GNG7 was correlated with poor prognosis of LUAD patients (a-b) Relationship between GNG7 expression and prognosis of LUAD patients in TCGA and GEO cohorts. (c) IHC results of LUAD tissues and normal tissues were scored according to the degree of staining according to 1–4. (d) Kaplan–Meier analysis was used to analyze correlation between GNG7 expression and prognosis of LUAD patients
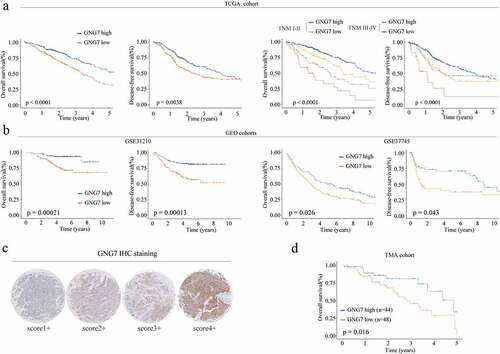
Figure 3. Overexpression of GNG7 inhibits proliferation, migration and invasion of LUAD cells (a-b) mRNA and protein levels of GNG7 were increased after transfection with GNG7-overexpressed plasmid in A549 and H1299 cells. (c) Cell viability of A549 and H1299 cells under treatment of GNG7 overexpressed plasmid and negative control in 1d,2d 3d and 4d. (d-e) GNG7 overexpression inhibited cell proliferation in LUAD cells, as demonstrated by colony formation and EDU assays, respectively. (f) The cell migration ability was measured by wound healing experiment in 0 h and 36 h after LUAD cell lines transfected with GNG7-overexpressed plasmid and negative control. (g) The invasion ability of A549 and H1299 cells with GNG7 overexpression were significantly suppressed according to cell invasion assay. Error bar represents the mean ± SD of three independent experiments. **p < 0.005.***p < 0.0005
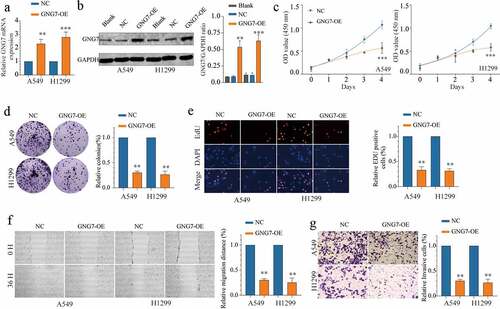
Figure 4. Overexpression of GNG7 inhibits tumorigenesis ability of LUAD cells in vivo (a-c) The in vivo effect of GNG7 was evaluated in xenograft mouse models bearing tumors originating from A549 cells via tumor volume and tumor weight; n = 5 per group. (d) Expression of GNG7 and Ki67 were detected through IHC staining between GNG7 overexpression and negative control groups in node mouse tumor tissues. **p < 0.005.***p < 0.0005
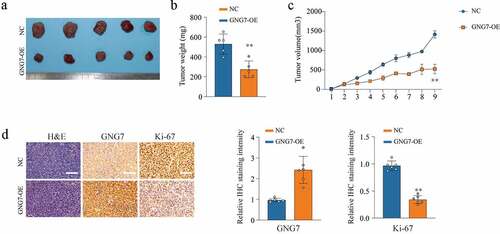
Figure 5. GNG7 was target regulated by miR-19b-3p in LUAD cells (a-c) Candidate miRNAs which upregulated in LUAD tissues and were complementary pairing with 3ʹUTR of GNG7 were showed, and GNG7 mRNA level was detected under condition of the candidate miRNAs mimics and negative control. (d) Relative luciferase activity between GNG7 wild type and mutant type based on a dual-luciferase reporter assay. (e, f) Protein and mRNA change of GNG7 in A549 and H1299 cells under treatment of miR-19b-3p mimics, inhibitor and negative control. (g-h) Expression difference of miR-19b-3p between LUAD tissues and normal tissues in RT-qPCR cohort and TCGA cohort. (mi) relationship between GNG7 expression and miR-19b-3p expression in LUAD tissues. Error bar represents the mean ± SD of three independent experiments. **p < 0.005.***p < 0.0005
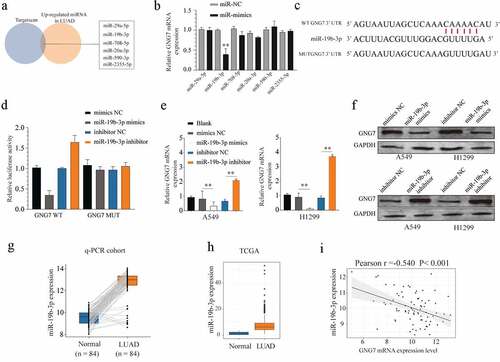
Figure 6. Overexpression of GNG7 could reverse the tumor-promoting effect of miR-19b-3p (a, b) showed GNG7 mRNA and protein expression under condition of miR-19b-3p mimics, miR-19b-3p mimics & GNG7 plasmid or negative control. (c) Cell viability of A549 and H1299 cells under treatment of miR-19b-3p mimics, miR-19b-3p mimics & GNG7 plasmid and negative control in 1d, 2d, 3d and 4d. (d) Colony formation ability of A549 and H1299 cells under treatment of miR-19b-3p mimics, miR-19b-3p mimics & GNG7 plasmid and negative control was compared in vitro, and the result was quantified. (e) Cell invasion ability of A549 and H1299 cells under treatment of miR-19b-3p mimics, miR-19b-3p mimics & GNG7 plasmid and negative control was detected by transwell assay, and the result was quantified. Error bar represents the mean ± SD of three independent experiments. **p < 0.005.***p < 0.0005, unpaired Student’s t test or one-way ANNOVA followed by multiple t test
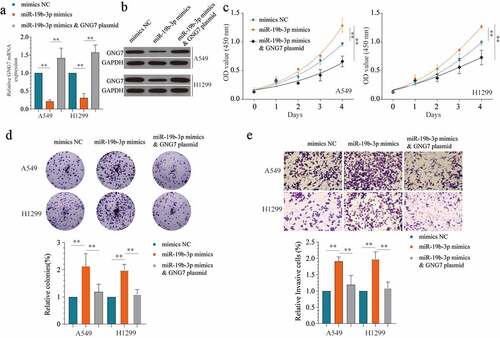
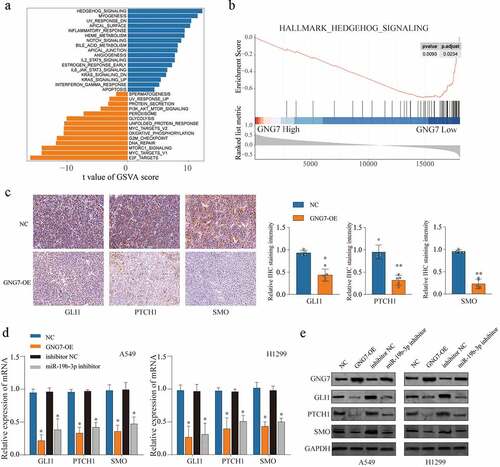
Figure 7. miR-19b-3p/GNG7 axis regulates progression of LUAD through hedgehog signaling the gene set enrichment analysis (a) and the gene set variation t analysis (b) was conducted in TCGA LUAD cohort between GNG7 high expression and GNG7 low expression, and indicated a significant correlation between GNG7 and Hedgehog pathway. (c) IHC staining for the change of SMO, GLI1 and PTCH1 levels after GNG7 overexpression. (d-e) Expression change of mRNA and protein levels of SMO, GLI1 and PTCH1 in A549 and H1299 cells which transfected with GNG7 overexpression plasmid or miR-19b-3p inhibitor, respectively. Error bar represents the mean ± SD of three independent experiments. **p < 0.0005