Figures & data
Figure 1. Elevated KCNQ1OT1 expression in CC. (a) RT-qPCR for determination of the expression differences of KCNQ1OT1 in CC tumor tissue specimens and adjacent non-tumor tissues. (b) RT-qPCR for determination of the expression differences of KCNQ1OT1 in CC cell lines (HeLa, SiHa, C33A) and normal human cervical epithelial cell line (End1/E6E7)
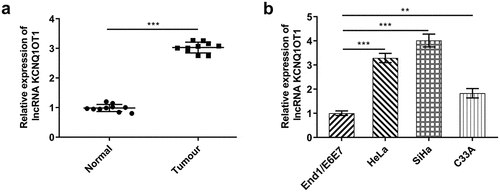
Figure 2. Silenced KCNQ1OT1 suppressed cell proliferation, migration and invasion in CC. (a) RT-qPCR validated interference efficacy in SiHa cells after transfection with KCNQ1OT1 knockdown lentiviral vectors. (b) CCK-8 assay examined the influence of KCNQ1OT1 silencing on CC cell proliferation. (c-d) Wound healing assay determined the influence of KCNQ1OT1 silencing on the migrative ability of CC cells. (e-f) Transwell assay detected the influence of KCNQ1OT1 silencing on the invasive ability of CC cells. (g) Western blotting analysis assessed the influence of KCNQ1OT1 silencing on the expressions of MMP2 and MMP9 in CC cells
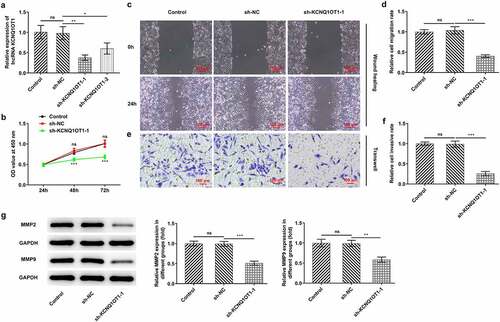
Figure 3. KCNQ1OT1 acted as a molecular sponge for miR-296-5p and negatively regulated miR-296-5p expression. (a) Bioinformatics analysis predicted the potential binding sites of miR-296-5p in KCNQ1OT1 sequence. (b) RT-qPCR validated overexpression efficacy in SiHa cells after transfection with miR-296-5p mimic. (c) Dual-luciferase reporter assay confirmed the binding relationship between KCNQ1OT1 and miR-296-5p. (d) RT-qPCR for determination of miR-296-5p level in SiHa cells after transfection with sh-KCNQ1OT1-1
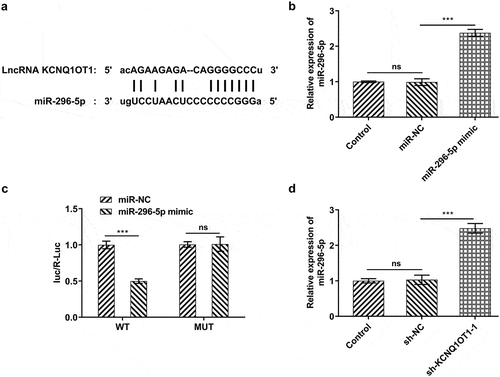
Figure 4. miR-296-5p was distinctly down-regulated in CC. (a) RT-qPCR for determination of the expression differences of miR-296-5p in CC tumor tissue specimens and adjacent non-tumor tissues. (b) RT-qPCR for determination of the expression differences of miR-296-5p in CC cell lines (HeLa, SiHa, C33A) and normal human cervical epithelial cell line (End1/E6E7)
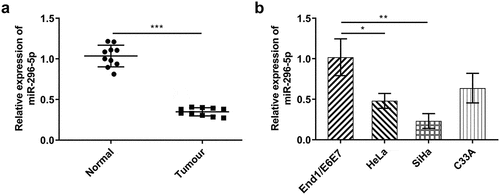
Figure 5. Upregulation of miR-296-5p counteracted the enhancing effects of overexpressed KCNQ1OT1 on the proliferative, migrative and invasive abilities of CC cells. (a) RT-qPCR for determination of KCNQ1OT1 level in SiHa cells after transfection with pcDNA-KCNQ1OT1 or co-transfection with miR-296-5p mimic. (b) CCK-8 assay examined the influence of miR-296-5p elevation on the enhanced proliferative ability of CC cells caused by KCNQ1OT1 overexpression. (c, d) Wound healing assay determined the influence of miR-296-5p elevation on the enhanced migrative ability of CC cells caused by KCNQ1OT1 overexpression. (e, f) Transwell assay detected the influence of miR-296-5p elevation on the enhanced invasive ability of CC cells caused by KCNQ1OT1 overexpression. (g) Western blotting analysis assessed the influence of miR-296-5p elevation on the increased expressions of MMP2 and MMP9 in CC cells caused by KCNQ1OT1 overexpression
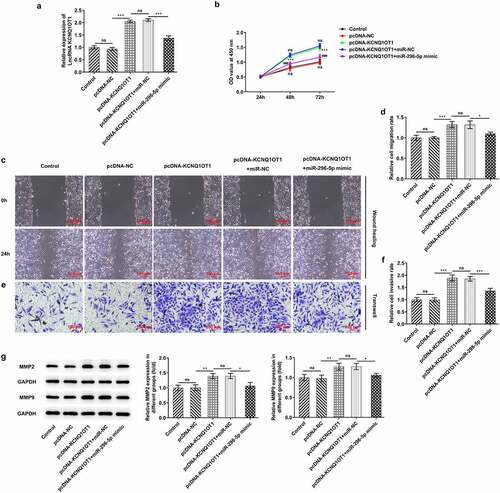
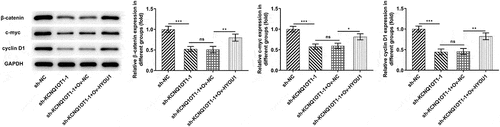
Figure 6. KCNQ1OT1 activated Wnt/β-catenin signaling pathway whereas miR-296-5p inactivated Wnt/β-catenin signaling pathway. Western blotting analysis examined the expressions of β-catenin, c-myc and cyclin D1 in SiHa cells after transfection with pcDNA-KCNQ1OT1 or co-transfection with miR-296-5p mimic
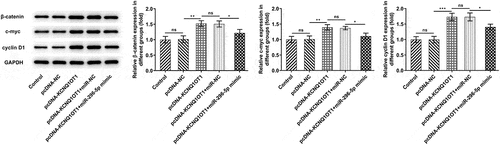
Figure 7. KCNQ1OT1 decoyed miR-296-5p to up-regulate its target gene HYOU1 in CC. (a) starBase predicted the complementary binding sites of miR-296-5p and HYOU1. (b) Dual-luciferase reporter assay confirmed the binding relationship between miR-296-5p and HYOU1. (c) RIP assay analyzed the potential binding between miR-296-5p and HYOU1. (d) Western blotting analysis examined the protein level of HYOU1 in CC cells after transfection with miR-296-5p mimic. (e) RT-qPCR detected the mRNA level of HYOU1 in CC cells after transfection with miR-296-5p mimic. (f) Western blotting analysis examined the protein level of HYOU1 in CC cells after transfection with pcDNA-KCNQ1OT1 or co-transfection with miR-296-5p mimic. (g) RT-qPCR detected the mRNA level of HYOU1 in CC cells after transfection with pcDNA-KCNQ1OT1 or co-transfection with miR-296-5p mimic
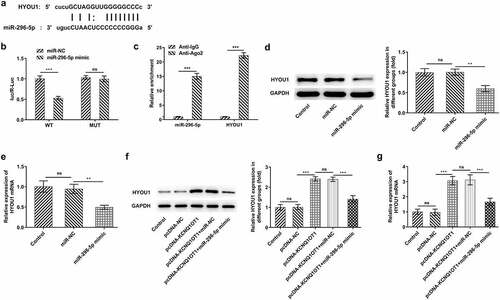
Figure 8. HYOU1 overexpression visibly abolished the suppressing effects of silenced KCNQ1OT1 on CC cell proliferation, migration and invasion. (a) RT-qPCR for determination of the expression differences of HYOU1 in CC cell lines (HeLa, SiHa, C33A) and normal human cervical epithelial cell line (End1/E6E7). (b) RT-qPCR verified the transfection efficiency in SiHa cells after transfection with Ov-HYOU1. (c) CCK-8 assay examined the influence of HYOU1 elevation on the suppressed proliferative ability of CC cells caused by silenced KCNQ1OT1. (d, e) Wound healing assay determined the influence of HYOU1 elevation on the suppressed migrative ability of CC cells caused by silenced KCNQ1OT1. (f, g) Transwell assay detected the influence of HYOU1 elevation on the suppressed invasive ability of CC cells caused by silenced KCNQ1OT1. (h) Western blotting analysis assessed the influence of HYOU1 elevation on the decreased expressions of MMP2 and MMP9 in CC cells caused by silenced KCNQ1OT1
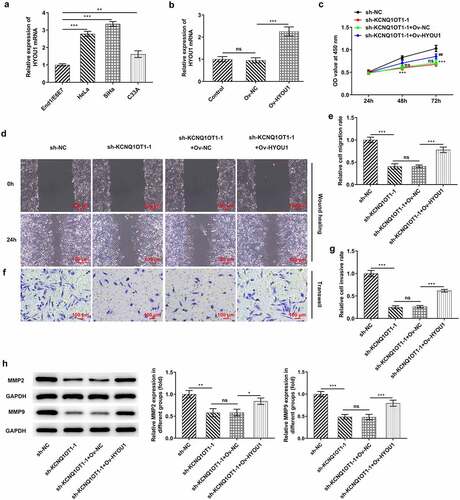
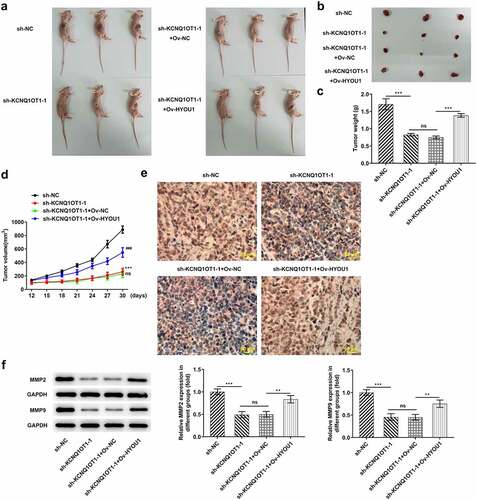
Figure 9. KCNQ1OT1 knockdown impeded tumor growth and metastasis by inhibiting HYOU1. (a) Representative morphologies of nude mice xenograft models. (b) Representative morphologies of xenograft tumors. (c) Weights of xenograft tumors. (d) Tumor volume curve. (e) Immunohistochemical staining examined Ki-67 expression in xenograft tumors. (f) Western blotting analysis detected expressions of MMP2 and MMP9 in xenograft tumors
Data availability
The analyzed data sets during the present study are available from the corresponding author on reasonable request.