Figures & data
Table 1. Characteristics of patients with ovarian cancers
Figure 1. Let-7b-5p is upregulated in OVCA and transfection efficiency of let-7b-5p inhibitor is promoted by USMB. (a) Let-7b-5p expression in OVCA tissues (N = 28) was revealed by RT-qPCR analysis. (b) The overall survival of OVCA patients with high or low level of let-7b-5p was presented using the method of Kaplan-Meier analysis. There were 14 patients with high let-7b-5p expression level and 14 patients with low let-7b-5p expression level, which was determined by the average value of let-7b-5p expression in OVCA patients. (c) The level of let-7b-5p in HOSEpiC, SKOV3 and CAOV3 cells was demonstrated by RT-qPCR. (d) The MBs were photographed by a microscope. (e) RT-qPCR was performed to evaluate let-7b-5p expression in SKOV3 and CAOV3 cells after transfection with NC inhibitor, let-7b-5p inhibitor and MB-let-7b-5p inhibitor. *P < 0.05, **P < 0.01, ***P < 0.001
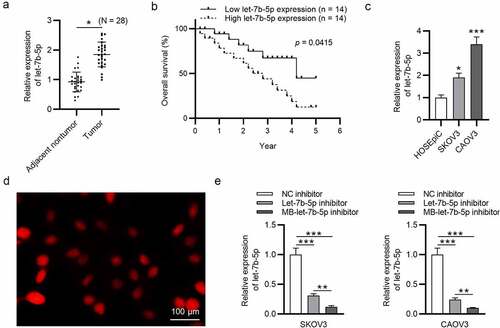
Figure 2. MB-let-7b-5p inhibitor has more suppressive effects on proliferation and stemness characteristics of OVCA cells than let-7b-5p inhibitor. SKOV3 and CAOV3 cells were divided into three groups and transfected with NC inhibitor, let-7b-5p inhibitor and MB-let-7b-5p inhibitor. (a) The CCK-8 assay was conducted to evaluate the cell viability. (b-c) Cell proliferative capacity was detected by colony formation assay. (d) Protein levels of stemness markers (OCT4, Nanog and SOX2) in each group were presented by western blot analysis. (e) The number of 3-D spheroids in each group was presented in sphere formation assay. *P < 0.05, **P < 0.01, ***P < 0.001
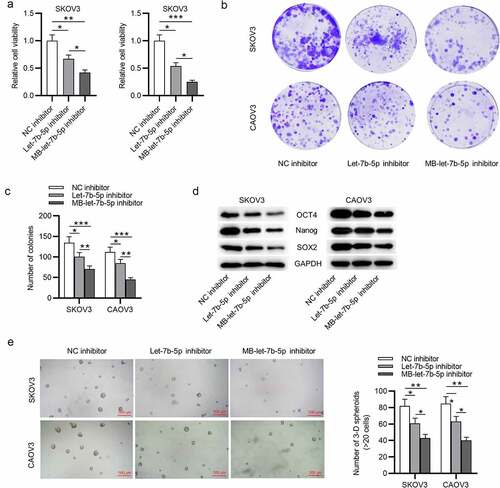
Figure 3. DDX19A is downregulated in OVCA. (a) The predicted mRNAs (GNG5, CHD4, NLK, DDX19A, PUDP and XKR8) targeted by let-7b-5p were provided by starBase (http://starbase.sysu.edu.cn/index.php) (filter criteria: Degradome Data: high stringency ≥ 3; Pan cancer: ≥ 10 cancer types; Program number = 5). (b) RT-qPCR was performed to detect the expression of GNG5, CHD4, NLK, DDX19A, PUDP and XKR8 in OVCA tissues (N = 28). (c) GEPIA predicted the expression of DDX19A in ovarian serous cystadenocarcinoma tissues (N = 426) and normal tissues (N = 88). (d) DDX19A level in SKOV3 and CAOV3 cells was measured by RT-qPCR. (e) Pearson correlation analysis presented the correlation between the expression of let-7b-5p and the level of DDX19A in OVCA tissues (N = 28). *P < 0.05, **P < 0.01, ***P < 0.001
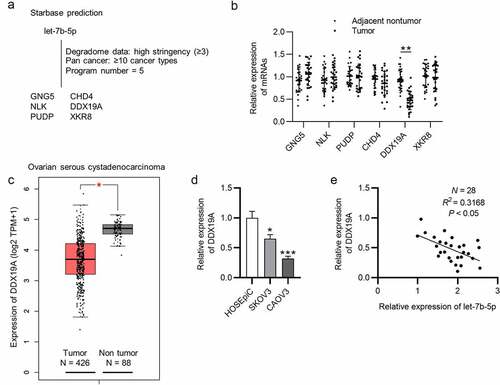
Figure 4. Let-7b-5p targets DDX19A 3ʹUTR and suppresses its expression. (a) DDX19A expression in OVCA cells after transfection with let-7b-5p inhibitor and MB-let-7b-5p inhibitor was presented by RT-qPCR. (b) Western blot presented the protein level of DDX19A in OVCA cells transfected with let-7b-5p inhibitor and MB-let-7b-5p inhibitor. (c) The binding sites between DDX19A 3ʹUTR and let-7b-5p were predicted by starBase. (d) Luciferase reporter assay presented the relative luciferase activity of pmirGLO-DDX19A 3ʹUTR after the transfection of let-7b-5p inhibitor and MB-let-7b-5p inhibitor. *P < 0.05, **P < 0.01, ***P < 0.001
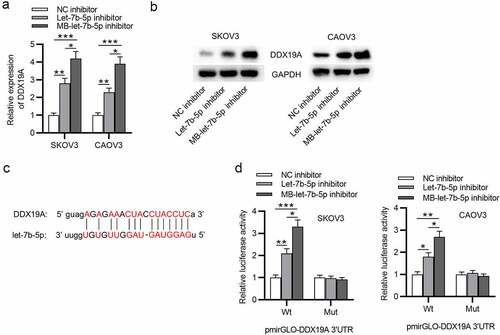
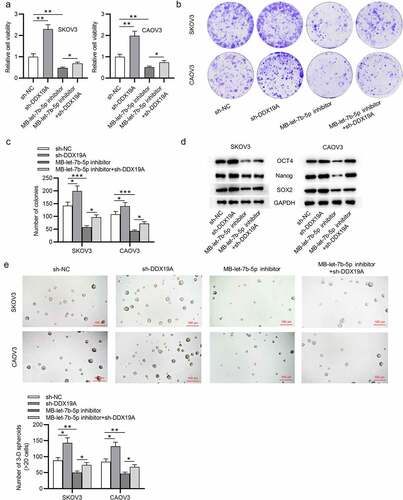
Figure 5. Silencing of DDX19A counteracts the effects of MB-let-7b-5p inhibitor on OVCA cells. SKOV3 and CAOV3 cells were divided into 4 groups: sh-NC, sh-DDX19A, MB-let-7b-5p inhibitor, MB-7b-5p inhibitor + sh-DDX19A. (a) Relative viability of cells in the 4 groups was assessed by the CCK-8 assay. (b-c) Colony formation assay was performed to evaluate the proliferation of OVCA cells. (d) The protein levels of stemness markers (OCT4, Nanog and SOX2) were measured by western blot analysis. (e) The number of 3-D spheroids formed by cells in the 4 groups was revealed by sphere formation assay. *P < 0.05