Figures & data
Table 1. Expression levels of candidate miRNAs
Figure 1. The expression of miR-877-3p in osteosarcoma. (a) In silico analysis using GEO database data GSE65071, including 20 human osteosarcoma tumor tissue samples and 15 control tissue samples. Differentially expressed miRNAs were plotted. (b) The expression of miR-877-3p in 71 samples of OS tissues and adjacent normal tissues were determined by real-time PCR. (c) The mRNA levels of miR-877-3p in OS tissues and adjacent normal tissues determined by real-time PCR. (d) overall survival of osteosarcoma patients based on miR-877-3p expression. (e) Expression level of miR-877-3p in osteosarcoma cells and normal osteoblast cell.
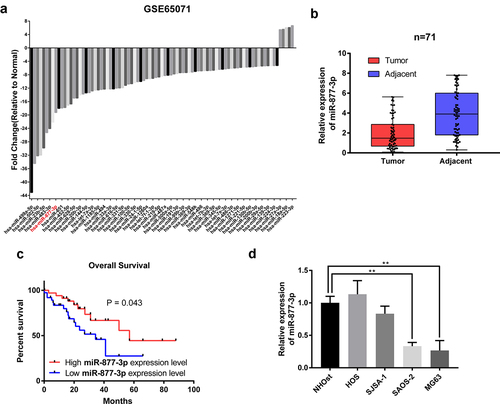
Figure 2. Effects of miR-877-3p on osteosarcoma proliferation. (a) After overexpression of miR-877-3p, MG63, and saos-2 cell proliferation was detected. Colony formation assay was performed to test the proliferation of (b) MG63 or (c) saos-2 cells. (d) Effect of miR-877-3p overexpression on cell cycle was measured by flow cytometry.
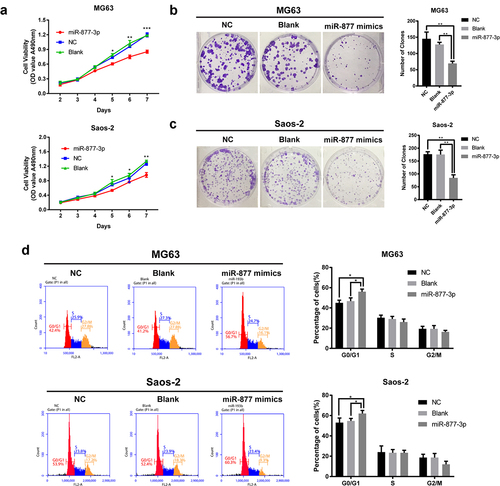
Figure 3. Effects of miR-877-3p on HUVECs. (a) Migration of transfected HUVECs in a scratch-wound assay 36 h after transfection. (b) Tube formation assay in HUVECs. (c) Heatmap of 84 angiogenesis genes in HUVECs.
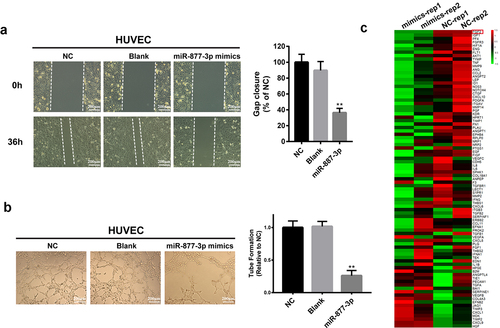
Figure 4. The relationship of miR-877-3p and FGF2 in osteosarcoma and HUVEC cells. After transfecting miR-877-3p mimics/control in MG63 and saos-2 cells. (a) The protein level of FGF2 was detected by Western blot analysis, (b) and the mRNA level of FGF2 was detected by real-time PCR. (c) The protein level and (d) the mRNA level of FGF2 were detected in HUVEC when transfected with miR-877-3p. (e) MiRDb predicted that miR-877-3p could target FGF2. (f) The interaction between miR-877-3p and FGF2 was examined by luciferase reporter assays.
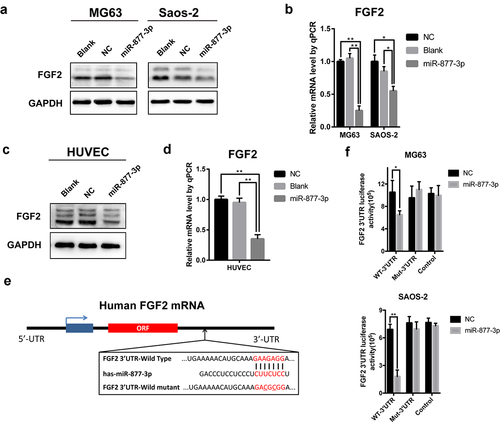
Figure 5. The effect of FGF2 overexpression in osteosarcoma and HUVEC cells. FGF2 was overexpressed in MG63 and saos-2 cells, cell proliferation was detected (a) by MTT assay and (b) colony formation assays. (c) HUVEC cells were transfected with control vector or miR-877-3p mimics only or both miR-877-3p mimics and FGF2 vectors, transfected HUVECs migration was detected in wound healing assay 36 h after transfection.
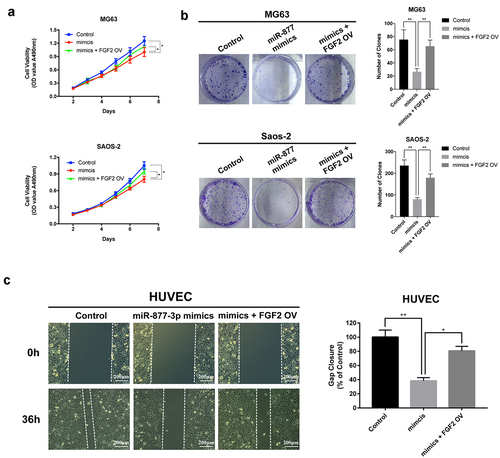
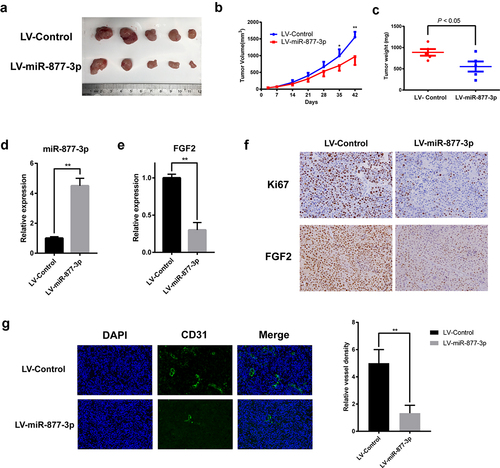
Figure 6. Effects of miR-877-3p on tumor growth and angiogenesis in mouse models. (a) MG63 tumors were harvested at the end of study, and (b) tumor volume and (c) tumor weight were measured. Expression level of miR-877-3p (d) and FGF2 (e) in tumor. (f) IHC staining of Ki67 and FGF2. (g) Immunofluorescence staining of CD31 for detection of blood vessels in MG63 tumor.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
The public data used in this study is available at http://www.ncbi.nlm.nih.gov/geo/