Figures & data
Table 1. Correlations of LINC00240 expression with clinicopathological features of gastric cancer
Figure 1. LINC00240 expression is increased both in GC tissues and GC cells. (a and b) qRT-PCR analysis showed the increased LINC00240 expression in GC tissues (n = 60). (a) and four GC cell lines (b) (*P < 0.05, **P < 0.01, ***P < 0.001). (c) Upregulation of LINC00240 expression in GC tissues from TCGA datasets. (d and e) High LINC00240 expression is associated with lower overall survival (d) and progression free survival (e) in GC patients
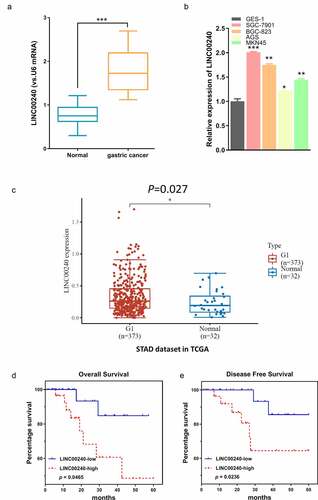
Figure 2. Knockdown of LINC00240 inhibits the proliferation and migration, and promotes cell apoptosis. (a) LINC00240 expression was quantified by qRT-PCR in GC cells (SGC-7901 and BGC-823) after siRNA transfection. (b) CCK-8 assay of the cell proliferation after knocking down LINC00240 in GC cells. (c) Colony formation assay, transwell migration (d) and invasion assay (e) in GC cell after siRNA transfection. (f) Flow cytometry analysis of apoptosis after LINC00240 knockdown (n = 3, **P < 0.01, ***P < 0.001)
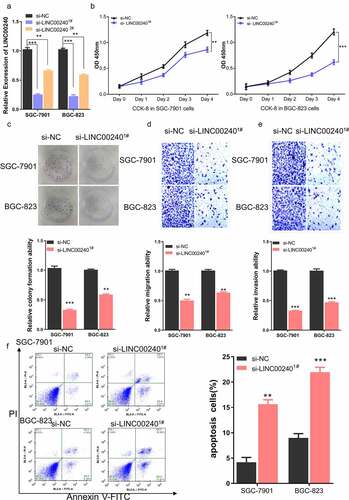
Figure 3. Evaluation the binding site between LINC00240 and miR-338-5p. (a) The binding site between LINC00240 and miR-338-5p based on bioinformatics analysis. (b) Increased expression level of miR-338-5p after LINC00240 knockdown detected by qRT-PCR. (c) Dual luciferase reporter assay using empty luciferase vector (luci-LINC00240-NC), wild type reporter with binding site (luci-LINC00240-WT), and the vector containing mutated binding sequence (luci-LINC00240-MUT) in the presence of miR-338-5p mimic. (n = 3, **P < 0.01). (d and e) Downregulation of miR-338-5p expression in GC tissue from TCGA datasets (d) and the GC tissues collected in this study (e). (f) Negative correlation between LINC00240 and miR-338-5p expression in GC tissues collected in this study (R2 = 0.5086, p < 0.001)
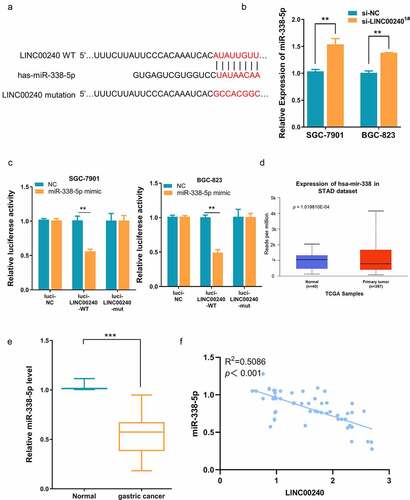
Figure 4. miR-338-5p inhibitor rescues si-LINC00240-induced inhibition on GC cell function. (a) qRT-PCR showed the downregulation of miR-338-5p by miR-338-5p inhibitor. (b-e) miR-338-5p inhibitor rescued si-LINC00240-induced inhibition on cell proliferation by CCK-8 assay (b), colony forming assay (c) and transwell migration and invasion assay (d and e). (f) miR-338-5p inhibitor reduced the level of apoptosis induced by si-LINC0024. (g) miR-338-5p mimic marginally downregulated LINC00240 level in GC cells. (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001)
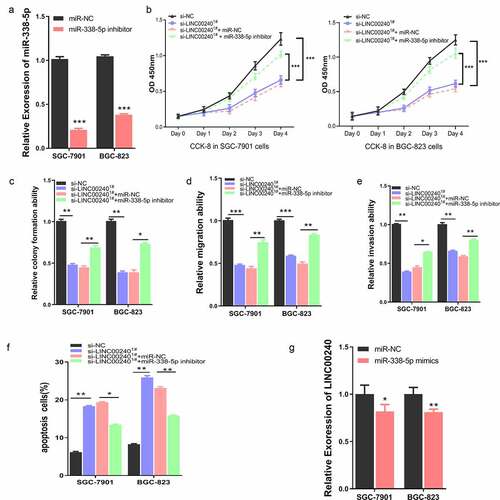
Figure 5. miR-338-5p targets METTL3 expression in GC cells. (a) The binding site between miR-338-5p and METTL3 3ʹ UTR from bioinformatics analysis. (b) Dual luciferase reporter assay using empty luciferase vector (luci-METTL3-NC), wild type reporter with binding site (luci-METTL3-WT), and the vector containing mutated binding sequence (luci-METTL3-MUT) in the presence of miR-338-5p mimic. (n = 3, ***P < 0.001). (c) Upregulation of METTL3 expression in GC tissue from TCGA datasets and (d) GC tissues collected in this study. (e) Negative correlation between METTL3 and miR-338-5p expression level in GC tissues collected in this study (R2 = 0.3456, p < 0.001). (f and g) Downregulation of METTL3 by miR-338-5p mimic as measured by qRT-PCR (f) and Western Blot assay (g) in SGC-7901 and BGC-823 cells (n = 3, **P < 0.01, ***P < 0.001)
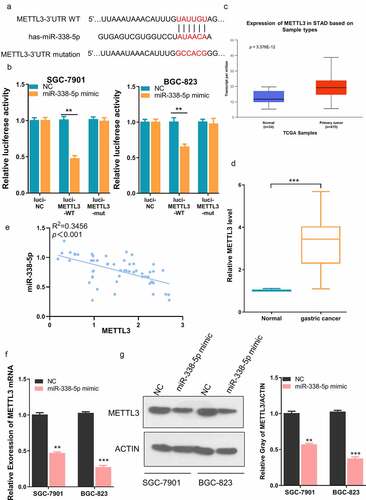
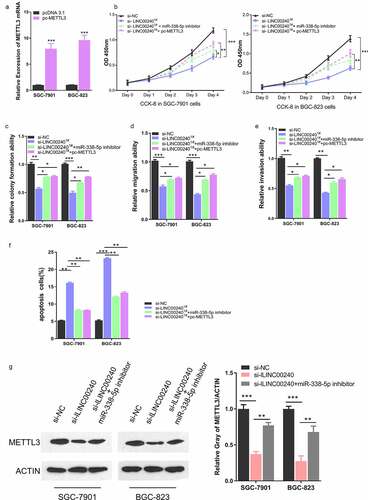
Figure 6. METTL3 overexpression rescues si-LINC00240-induced inhibition on GC cell function. (a) Overexpression of METTL3 after the transfection of pc-METTL3 expression vector. (b-f) GC cells were transfected with si-NC, si-lLINC00240, si-lLINC00240+ miR-338-5p inhibitor, or si-lLINC00240+ pc-METTL3. (b) CCK-8 Cell proliferation, (c) colony formation, (d) cell migration, (e) cell invasion, and (f) apoptosis assays were performed in GC cells 48 hours after the transfection. (g) Expression of METTL3 was measured by Western Blot after transfecting with si-lLINC00240 or si-lLINC00240+ miR-338-5p inhibitor. (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001)