Figures & data
Table 1. The primer sequences used for qRT-PCR
Figure 1. Exosomes mediated the transport of miR-4488 and miR-1273 g-5p in TGF-β2-treated ARPE-19 cells. (a-b) The expression of miR-4488 and miR-1273 g-5p in ARPE-19 cells treated with or without TGF-β2 was measured by qRT-PCR. (c) The morphology of exosomes from TGF-β2-treated ARPE-19 cells was analyzed using TEM. (d) The expression of exosome markers (CD9, CD63 and CD81) was detected by WB analysis. (e) The size of exosomes was analyzed by NTA. (f) Donor ARPE-19 cells were transfected with miR-4488 mimic or miR-1273 g-5p mimic followed by treated with TGF-β2. The expression of miR-4488 and miR-1273 g-5p was detected by qRT-PCR. (g) Exosomes were isolated from donor ARPE-19 cells transfected with miR-4488 mimic or miR-1273 g-5p mimic followed by treated with TGF-β2. The expression of miR-4488 and miR-1273 g-5p in exosomes was determined by qRT-PCR. (h) Recipient ARPE-19 cells were treated with exosomes followed by treated with TGF-β2. The expression of miR-4488 and miR-1273 g-5p were detected by qRT-PCR. **P < 0.01, ***P < 0.001
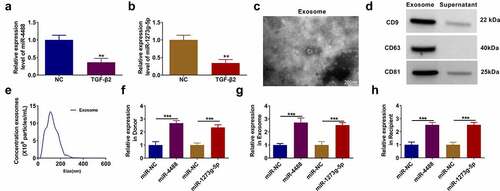
Figure 2. Exosome miR-4488 inhibited the EMT, migration and invasion of TGF-β2-induced ARPE-19 cells. ARPE-19 cells were treated with exosomes (Exo-miR-NC or Exo-miR-4488) followed by treated with or without TGF-β2. Non treated cells were used as normal control (NC). (a) The expression of miR-4488 was measured by qRT-PCR. (b) The protein levels of ZO-1, occludin, E-cadherin and α-SMA were detected by WB analysis. (c-d) The migration and invasion abilities were measured by transwell assay. (e) Wound healing assay was used to assess the migration of cells. (f-g) The apoptosis rate of cells was determined by flow cytometry. (h) The protein levels of Bcl-2, Bax and cleaved caspase 3 were assessed by WB analysis. **P < 0.01, ***P < 0.001, ****P < 0.0001
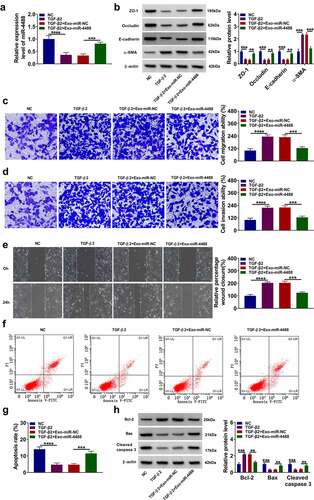
Figure 3. MiR-4488 directly targeted ABCA4. (a) The binding sites and mutant sites between ABCA4 3ʹUTR and miR-4488 were shown. Dual-luciferase reporter assay (b) and RIP assay (c) were used to confirm the interaction between ABCA4 and miR-4488. (d-e) The mRNA and protein levels of ABCA4 in ARPE-19 cells treated with or without TGF-β2 was determined by qRT-PCR and WB analysis. (f) ARPE-19 cells were transfected with or without pcDNA or pcDNA ABCA4 overexpression vector, and then treated with or without Exo-miR-4488 or Exo-miR-NC followed by treated with TGF-β2. The protein level of ABCA4 was detected by WB analysis. **P < 0.01, ***P < 0.001
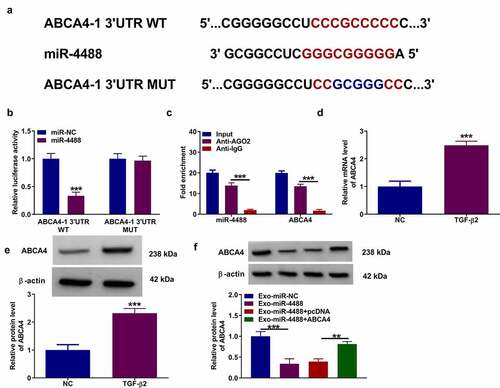
Figure 4. ABCA4 reversed the effect of exosomal miR-4488 on the EMT, migration and invasion of TGF-β2-induced ARPE-19 cells. ARPE-19 cells were transfected with or without pcDNA or pcDNA ABCA4 overexpression vector and then treated with or without Exo-miR-4488 or Exo-miR-NC followed by treated with TGF-β2. (a) WB analysis was used to detect the protein levels of ZO-1, occludin, E-cadherin and α-SMA. (b-c) Transwell assay was performed to measure cell migration and invasion abilities. (d) The migration of cells was evaluated using wound healing assay. (e-f) The apoptosis rate of cells was assessed using flow cytometry. (g) WB analysis was utilized to determine the protein levels of Bcl-2, Bax and cleaved caspase 3. *P < 0.05, **P < 0.01, ***P < 0.001
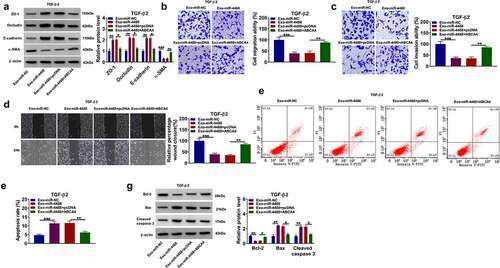
Figure 5. Exosomal miR-1273 g-5p had an inhibition on the EMT, migration and invasion of TGF-β2-induced ARPE-19 cells. ARPE-19 cells were treated with exosomes (Exo-miR-NC or Exo-miR-1273 g-5p) followed by treated with or without TGF-β2. Non treated cells were used as normal control (NC). (a) The miR-1273 g-5p expression was detected by qRT-PCR. (b) The protein levels of ZO-1, occludin, E-cadherin and α-SMA were determined using WB analysis. (c-d) Transwell assay was utilized to test cell migration and invasion abilities. (e) Wound healing assay was performed to measure cell migration. (f-g) Cell apoptosis rate was evaluated by flow cytometry. (h) The protein levels of Bcl-2, Bax and cleaved caspase 3 were analyzed by WB analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
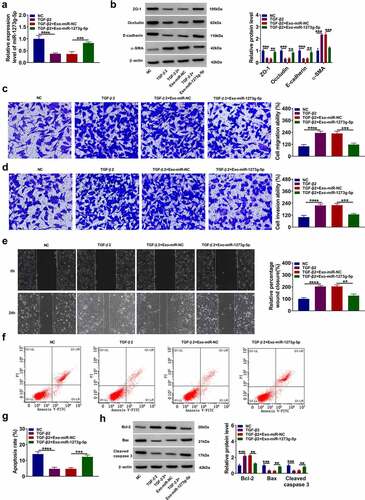
Figure 6. ABCA4 also could be targeted by miR-1273 g-5p. (a) The binding sites and mutant sites between ABCA4 3ʹUTR and miR-1273 g-5p were shown. Dual-luciferase reporter assay (b) and RIP assay (c) were used to confirm the interaction between ABCA4 and miR-1273 g-5p. (d) ARPE-19 cells were transfected with or without pcDNA or pcDNA ABCA4 overexpression vector and then treated with or without Exo-miR-1273 g-5p or Exo-miR-NC followed by treated with TGF-β2. WB analysis was used to measure the protein level of ABCA4. ***P < 0.001, ****P < 0.0001
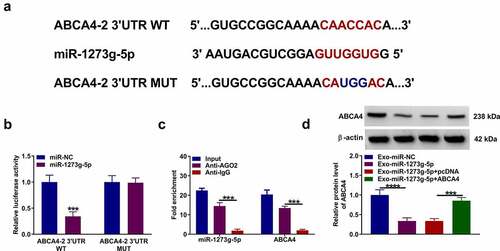
Figure 7. Exosomal miR-1273 g-5p regulated the EMT, migration, invasion and apoptosis of TGF-β2-induced ARPE-19 cells by targeting ABCA4. ARPE-19 cells were transfected with or without pcDNA or pcDNA ABCA4 overexpression vector and then treated with or without Exo-miR-1273 g-5p or Exo-miR-NC followed by treated with TGF-β2. (a) The protein levels of ZO-1, occludin, E-cadherin and α-SMA were measured by WB analysis. (b-c) Transwell assay was performed to detect cell migration and invasion abilities. (d) Wound healing assay was used to detect the migration of cells. (e-f) Cell apoptosis rate was analyzed using flow cytometry. (g) WB analysis was used to measure the protein levels of Bcl-2, Bax and cleaved caspase 3. *P < 0.05, **P < 0.01, ***P < 0.001
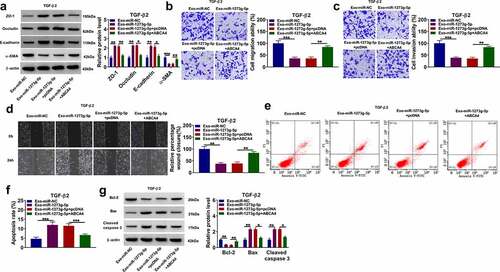
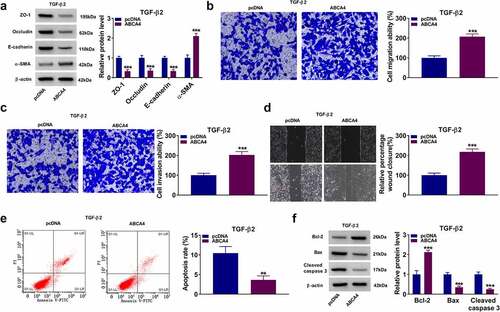
Figure 8. ABCA4 overexpression promoted EMT, migration and invasion in TGF-β2-induced ARPE-19 cells. ARPE-19 cells were transfected with pcDNA or pcDNA ABCA4 overexpression vector followed by treated with TGF-β2. (a) WB analysis was performed to detect the protein levels of ZO-1, occludin, E-cadherin and α-SMA. (b-c) Cell migration and invasion abilities were determined by transwell assay. (d) Wound healing assay was utilized for measuring the migration of cells. (e) Flow cytometry was used to examine cell apoptosis rate. (f) WB analysis was performed to analyze the protein levels of Bcl-2, Bax and cleaved caspase 3. **P < 0.01, ***P < 0.001