Figures & data
Figure 1. General COVID-19 diagnostic workflow using molecular testing (NAAT, iNAAT and immunoassay-based detection). (1) Sample collection methods; (2) Types of samples; (3) Sample processing or pre-treatment; (4) Test reaction and result reading. The methods illustrated are the most commonly used for COVID-19 diagnosis and the alternatives in each step are mostly interchangeable, except that blood samples are not used for NAAT and iNAAT techniques, and extracted RNA is not used for immunoassay detection. The image was created with BioRender.com
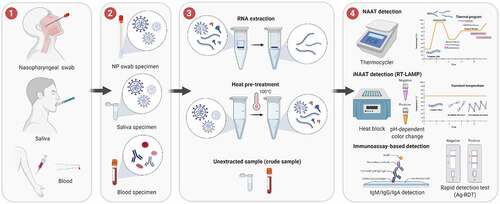
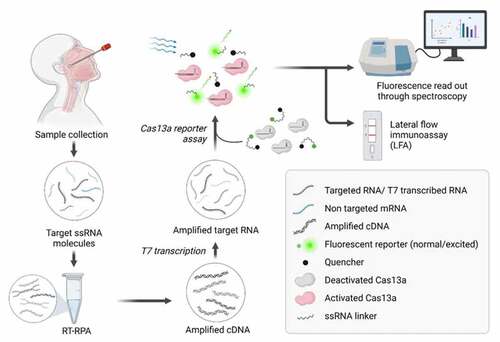
Figure 2. CRISPR-Cas13a based detection of SARS-CoV-2. Nasopharyngeal and oropharyngeal specimens are collected via sterile swabs. The collected sample is then diluted in an appropriate buffer, followed by a few heating steps. Sample heating steps release ssRNA from the virus and facilitate the deactivation of nuclease if any is present in the sample. Following the heating step, the viral RNA is subjected to RT-RPA for the amplification of target sequences in the form of cDNAs, which are in turn transcribed by T7 RNA polymerase. The accumulated amplification products of the targeted RNA sequence are provided for Cas13a-based detection assay. Cas13a recognizes T7-transcribed RNA sequences if appropriate guide RNA (gRNA) is presented. This leads to the activation of Cas13a and displays its nonspecific RNAase activity, resulting in the nonspecific cleavage of the fluorophore-ssRNA-quencher complex. The florescence emitted by the fluorophore can be quantified via spectroscopy, indicating the concentration of the ssRNA template. Alternatively, the cleaved reporter molecule can be detected via paper LFA. The image was created with BioRender.com