Figures & data
Figure 1. The overall protocol of this study
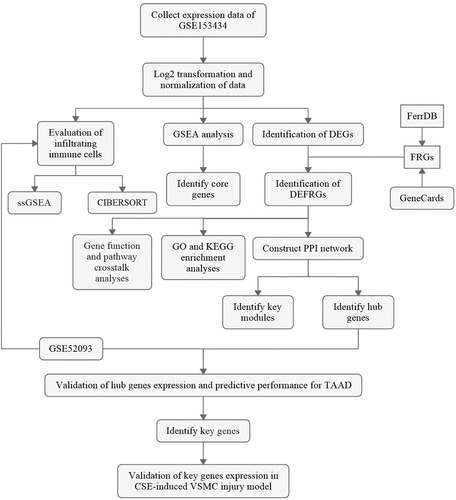
Figure 2. GSEA and identification of differentially expressed genes
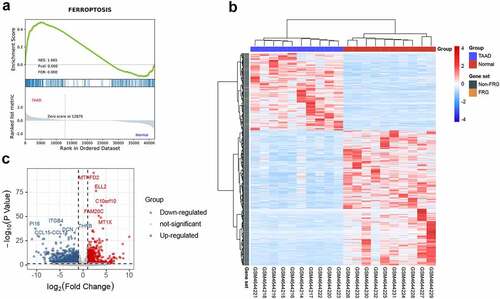
Figure 3. Identification of differentially expressed ferroptosis-related genes
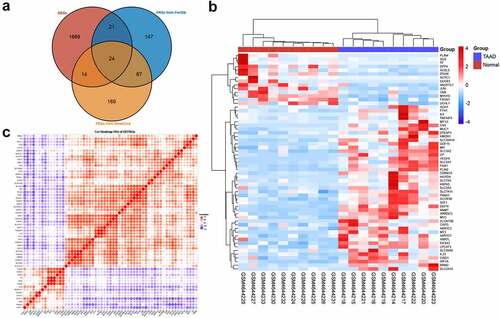
Figure 4. GO and KEGG enrichment analyses of differentially expressed ferroptosis-related genes
Figure 5. PPI network and identification of key modules and hub genes
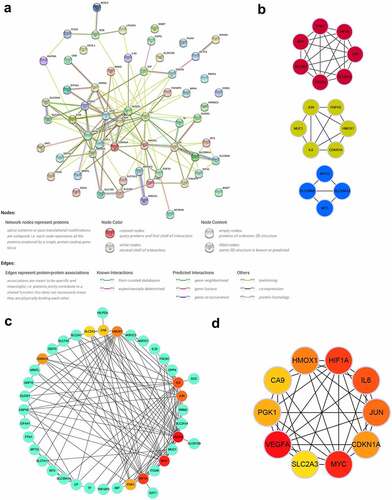
Figure 6. Validation of hub genes in GSE52093 and CSE-induced VSMC injury model
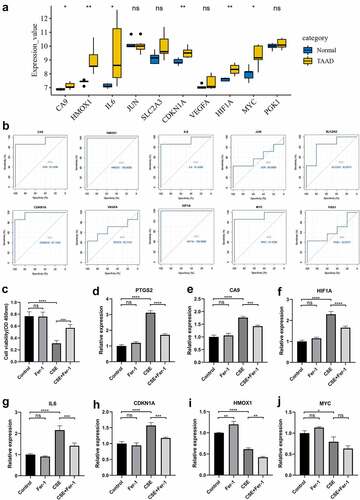
Figure 7. Immune infiltration analyses by the CIBERSORT algorithm in GSE153434 and GSE52093
Supplemental Material
Download Zip (796.6 KB)Data availability
Publicly available datasets were analyzed in this study. The data used to support the results of this study are available from the online website as mentioned above.
