Figures & data
Figure 1. The flow chart for the methods of the bio-information analysis, which clearly showed the process of the identification of prognostic lncRNA signature in HNSCC
Figure 2. LASSO regression analysis was used to select DElncRNAS related to prognosis of HNSCC. (a) Forest plots showing the relationship between different lncRNA subsets and OS in the training queue. Unadjusted HRs are 95% CIs. (b) The box diagram shows the differential gene expression of model lncRNA in TCGA database. ***P < 0.001, **P < 0.01, and *P < 0.05
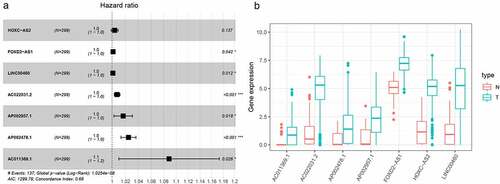
Figure 3. Validation of lncRNA signaling in predicting the prognosis of HNSCC in the training group. (a) Risk score distribution of patients in the training group. (b) Survival status distribution of high- and low-risk patients. (c) Kaplan-Meier OS curves were constructed for low- and high-risk HNSCC patients. (d) Comparison of the predictive OS ability of the risk score model with other clinical parameters using ROC curve analysis for HNSCC patients. (e) Relationship between risk factors and HNSCC OS (univariate Cox regression analysis). (f) Relationship between risk factors and HNSCC OS (multivariate Cox regression analysis)
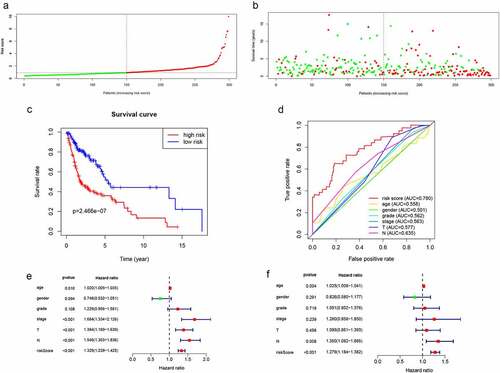
Figure 4. Validation of lncRNA signaling in predicting the prognosis of HNSCC in the validation group. (a-d) The results of lncRNA signature prediction of HNSCC outcomes in the validation group were consistent with the results of the training cohort(). (e) Relationship between risk factors and HNSCC OS in the validation group (univariate Cox regression analysis). (f) Relationship between risk factors and HNSCC OS in the validation group (multivariate Cox regression analysis)
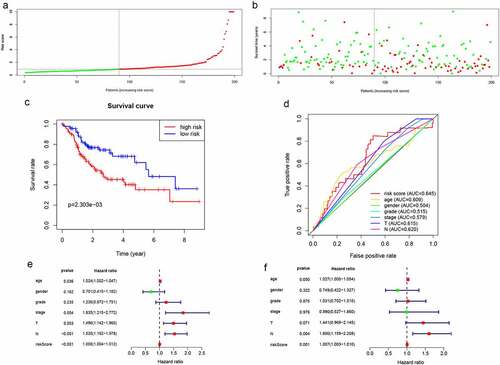
Figure 5. Levels of FOXD2-AS1 is significantly increased in HNSCC tissues and associated with poor prognosis. (a) The expression of FOXD2-AS1 in HNSCC tissues (n = 501) compared with normal tissues (n = 44) was analyzed using TCGA data. (b) FOXD2-AS1 level was verified in 44 paired HNSCC carcinoma and paracarcinoma tissues from the TCGA database. (c) Kaplan-meier plotter was used to determine the relationship between FOXD2-AS1 levels and overall survival in HNSCC patients. (d-e) Univariate and multivariate COX regression analysis showed that FOXD2-AS1 level was an independent predictor of HNSCC. (f) GSEA describes the biological pathways associated with FOXD2-AS1
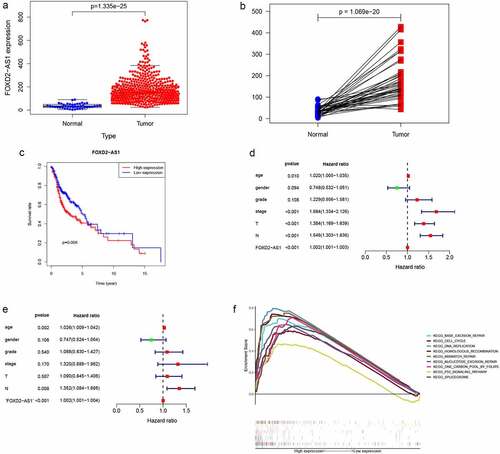
Figure 6. Clinicopathologic features of FOXD2-AS1 expression. (a-d) FOXD2-AS1 expression significantly increased in advanced cases
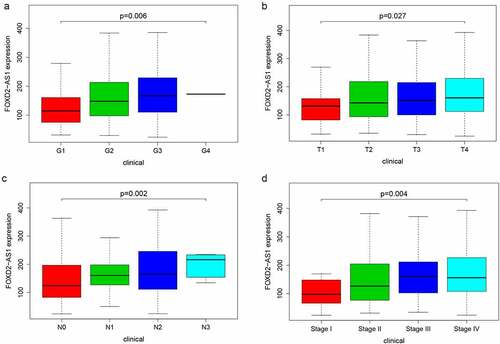
Figure 7. The expression and biological characteristics of FOXD2-AS1 in HNSCC in vitro study. (a) FOXD2-AS1 was remarkably increased in HNSCC cell lines. (b) Transfection efficiency was verified after lentivirus transfection of FOXD2-AS1 or negative control shRNA. (c) Expression and localization of FOXD2-AS1 in HOK, TSCCA, CAL-27 and FADU cell lines
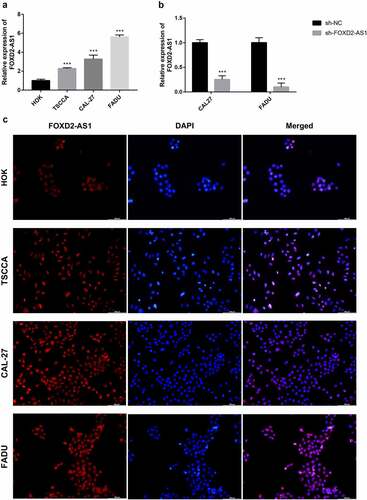
Figure 8. The expression and biological characteristics of FOXD2-AS1 in HNSCC in vitro study. (a-b) The viability of HNSCC cells was assessed by CCK-8 assay at 0, 24, 48 and 72 h after transfection. (c) Images were recorded 0 and 48 hours after the cell scratch experiment. (d-e) Transwell assay was used to detect the invasion and migration of HNSCC cells. ***P < 0.001, **P < 0.01, and *P < 0.05
Supplemental Material
Download Zip (4.6 MB)Availability of data and materials
This study obtained open data from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/).
