Figures & data
Table 1. The correlation between clinicopathological features and expression of circ_0048764 in BC patients
Figure 1. Circ_0048764 is up-regulated in BC tissues and cells (a) the GSE165884 dataset was analyzed by the GEO2R online tool to show the expression changes of circRNAs in BC tissues and normal tissues, with red representing the upregulated circRNAs and blue representing the downregulated circRNAs. (b) A heat map showed the differential expression of circRNAs in BC tissues and normal tissues. (c and d) qRT-PCR was used to detect the expression of circ_0048764 in BC tissues (c) and cell lines (d). Nucleocytoplasmic separation assay was used to detect the subcellular localization of circ_0048764 in SKBR3 and MCF7 cells. **P < 0.01, and ***P < 0.001.
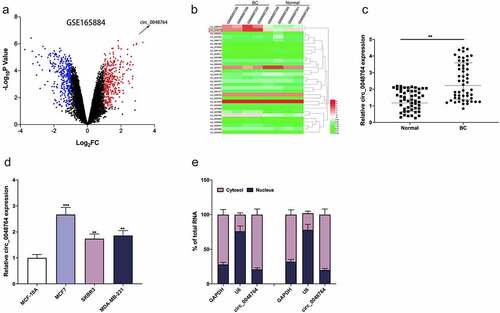
Figure 2. Circ0048764 promotes BC cells’ proliferation, migration and invasion and inhibits apoptosis (a) pcDNA-NC or pcDNA-circ_0048764 was transfected into SKBR3 cells and si-NC, si-circ0048764-1 and circ_0048764-2 were transfected into MCF7 cells, and the expression of circ_0048764 in SKBR3 and MCF7 cells after transfection was detected by qRT-PCR. (b and c) CCK-8 and EdU assays were used to detect the effects of overexpression or knockdown of circ_0048764 on the proliferation of SKBR3 and MCF7 cells. (d) transwell assay was used to detect the effects of overexpression or knockdown of circ_0048764 on SKBR3 and MCF7 cells’ migration and invasion. (e) flow cytometry was used to detect the effect of overexpression or knockdown of circ_0048764 on the apoptosis of SKBR3 and MCF7 cells. ** P < 0.01, and ***P < 0.001.
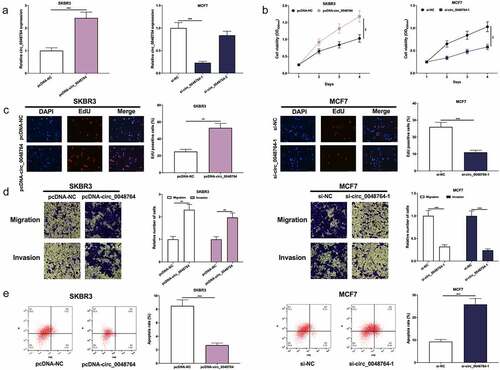
Figure 3. Circ_0048764 adsorbs miR-1296-5p (a) The potential binding site between circ_0048764 and miR-1296-5p was predicted by the Circinteractome database. (b) The interaction between circ_0048764 and miR-1296-5p was detected with a dual-luciferase reporter gene assay with HEK-293 T cells . (c) RIP assay was performed to detect the enrichment of circ_0048764 and miR-1296-5p in the immunoprecipitate of Ago2 group or IgG group. (d) qRT-PCR was used to detect the expression of miR-1296-5p in SKBR3 and MCF7 cells with circ_0048764 overexpression or knockdown. e and f. The expression of miR-1296-5p in BC tissues (e) and cell lines (f) was detected by qRT-PCR. (g) Pearson’s correlation analysis was performed to detect the correlation between circ_0048764 expression and miR-1296-5p expression in BC tissues. * P < 0.05, ** P < 0.01, and *** P < 0.001.
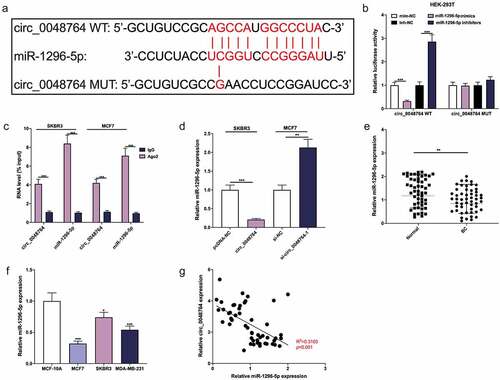
Figure 4. Circ_0048764 promotes BC cell proliferation, migration and invasion, and inhibits apoptosis by targeting miR-1296-5p (a) pcDNA-NC, pcDNA-circ_0048764 and pcDNA-circ_0048764 + miR-1296-5p mimics were transfected into SKBR3 cells, respectively; si-NC, si-circ_0048764-1 and si-circ_0048764-1 + miR-1296-5p inhibitors were transfected into MCF7 cells, respectively, and the expression of miR-1296-5p was detected by qRT-PCR. (b and c) CCK-8 and EdU assays were used to detect SKBR3 or MCF7 cells’ proliferation after transfection. (d) transwell assay was used to detect SKBR3 or MCF7 cells’ migration and invasion after transfection. (e) flow cytometry was used to detect the apoptosis of SKBR3 or MCF7 cells after transfection. ** P < 0.01, and ***P < 0.001.
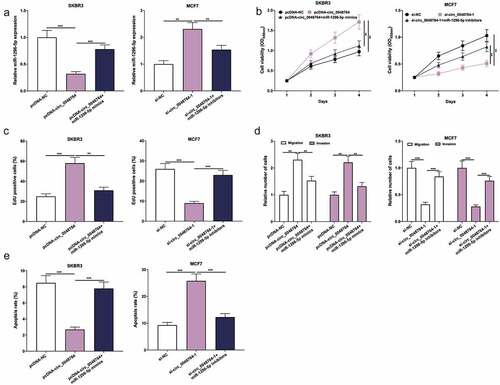
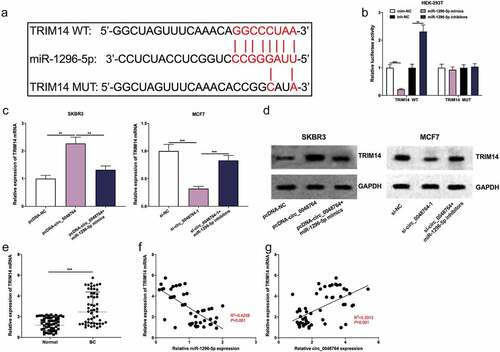
Figure 5. Circ_0048764/miR-1296-5p axis regulates BC progression by regulating the expression of TRIM14 (a)The potential binding site between TRIM14 mRNA 3′ UTR and miR-1296-5p were predicted by targetscan database. (b) dual-luciferase reporter gene assay was used to examine the interaction of TRIM14 mRNA 3’UTR and miR-1296-5p in HEK-293 T cells. (c and d) the levels of TRIM14 mRNA and protein in SKBR3 cells transfected with pcDNA-NC, pcDNA-circ_0048764 and pcDNA-circ_0048764 + miR-1296-5p mimics, or MCF7 cells transfected with si-NC, si-circ_0048764-1 and si-circ_0048764-1 + miR-1296-5p inhibitors were detected by qRT-PCR and western blot assay, respectively. (e) the expression of TRIM14 mRNA in BC tissues and normal tissues was detected by qRT-PCR. (f and g) Pearson’s correlation analysis was used to detect the correlation between TRIM14 mRNA expression and miR-1296-5p expression or circ_0048764 expression in BC tissues, respectively. ** P < 0.01, and ***P < 0.001.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
