Figures & data
Figure 1. LATS2 mRNA expression in CRC tissues and cell lines. (a) LATS2 mRNA expression was significantly decreased in CRC tissues compared with matched adjacent normal tissues. (b-c) In the TCGA database, LATS2 mRNA expression was markedly lower in COAD and READ tissues compared with normal tissues. (d) LATS2 expression was decreased in CRC cell lines compared with a normal intestinal epithelial cell (NCM460). Data were expressed as mean ± SD, *P < 0.05; **P < 0.01; ***P < 0.001
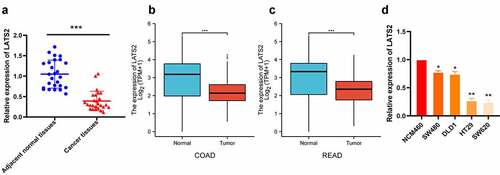
Table 1. LATS2 expression in colorectal benign and malignant tissues
Figure 2. Representative patterns of LATS2 protein expression in colorectal benign and malignant tissues on TMA sections. (a–b) Colorectal cancer with low or no LATS2 expression. (c–d) Colorectal cancer with high LATS2 expression. (e–f) High-grade intraepithelial neoplasia with low LATS2 expression. (g–h) Low-grade intraepithelial neoplasia with low LATS2 expression. (i–j) Normal surgical margin of colorectal cancer with high LATS2 expression
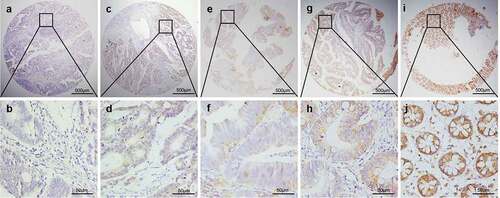
Table 2. Associations of LATS2 protein expression with clinicopathological characteristics in colorectal cancer patients
Table 3. Univariate and multivariate analyses of prognostic factors for overall survival in colorectal cancer patients
Figure 3. Survival analysis of CRC patients by the Kaplan–Meier method. (a) Overall survival in patients with high LATS2 expression was significantly higher than that in patients with low LATS2 expression. (b) Overall survival in patients with stage II or stage III–IV CRC was significantly lower than that in patients with stage 0–I CRC
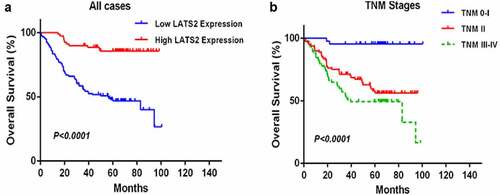
Figure 4. Notable immune-related signaling pathways in the high LATS2 expression group. (a) LATS2-activated signaling pathways. (b) Chemokine signaling pathways. (c) Cytokine-cytokine receptor interaction signaling pathways. (d) JAK-STAT signaling pathways. (e) Intestinal immune network for IgA production signaling pathways
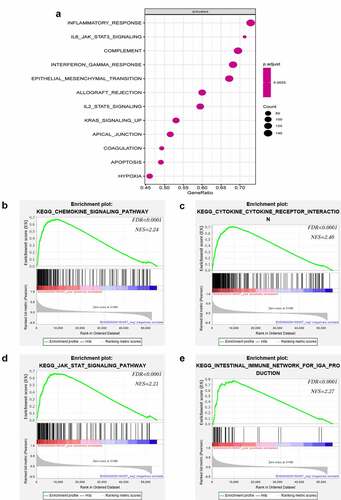
Table 4. Correlations between LATS2 expression and marker genes for tumor-infiltrating immune cells using the TIMER database
Figure 5. Correlations of LATS2 expression with tumor microenvironment and immune infiltration levels in CRC. (a) Positive correlation between LATS2 expression and immune score in CRC. (b) LATS2 was positively correlated with tumor-infiltrating immune cells in COAD by TIMER. (c) LATS2 was positively correlated with tumor-infiltrating immune cells in READ by TIMER. (d) LATS2 was positively correlated with tumor-infiltrating immune cells in COAD by ssGSEA. (e) LATS2 was positively correlated with tumor-infiltrating immune cells in READ by ssGSEA
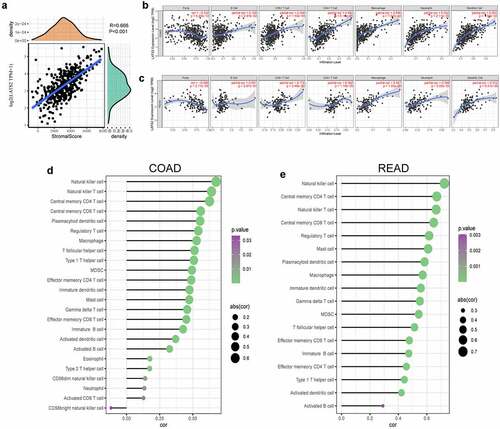
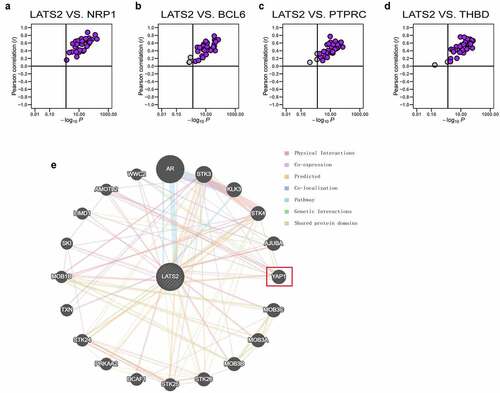
Figure 6. Correlations of LATS2 expression with NRP1, BCL6, PTPRC, and THBD expression in cancer samples and analysis of the PPI network for LATS2. (a–d) LATS2 was positively associated with NRP1, BCL6, PTPRC, and THBD expression in almost all cancers examined, based on data from the TCGA. (e) Protein-protein network view in the GeneMANIA dataset showing the interaction networks of LATS2
Supplemental Material
Download Zip (585.8 KB)Data availability statement
The gene expression data of CRC and normal tissues were extracted from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga). All the data will be provided on reasonable request from the corresponding author.
