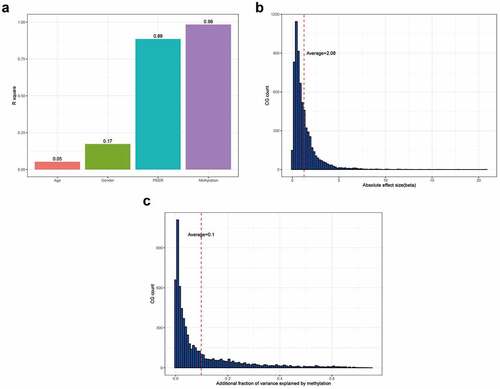
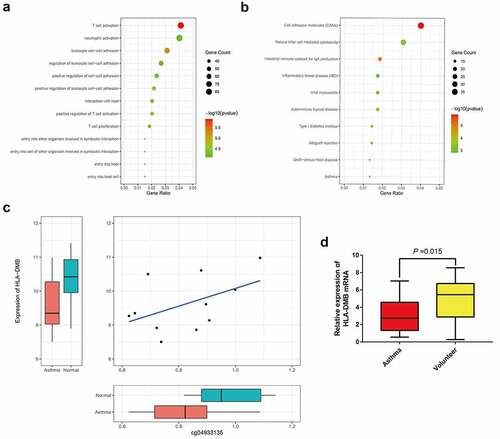
Figures & data
Table 1. Primer for MSP-PCR
Figure 1. DNA methylation in asthma. (a) The distribution of differential methylation sites on chromosomes; (b) Frequency of hypermethylation and hypomethylation on each chromosome; (c) Clustering heat map of methylation difference sites
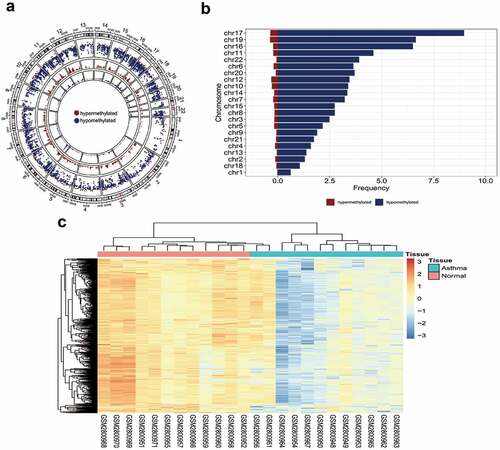
Table 2. Distribution of methylation on each chromosome
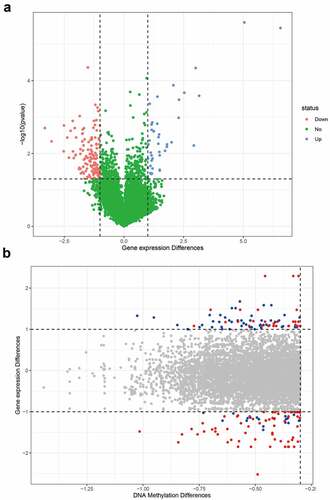
Figure 2. Differential genes in asthma. (a) Volcano map of differential genes; (b) The relationship between DNA methylation differences and expression of differential genes. The blue dots indicate that the fold change of gene expression was greater than 2, and the average difference of methylation sites was greater than 0.3. The red dots indicate the sites with statistical differences
Supplemental Material
Download JPEG Image (1.2 MB)Data availability statement
The data used to support the findings of this study are available from the corresponding author ([email protected]) upon request.