Figures & data
Table 1. Primer sequences used in this study
Figure 1. Circ_0000423 expression is significantly up-regulated in GC tissues and cell lines
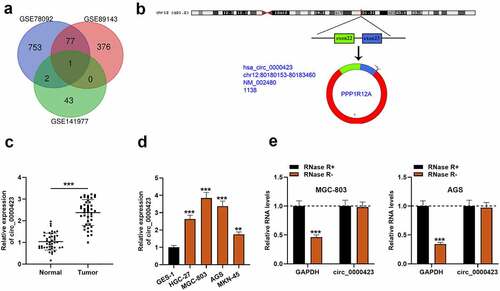
Figure 2. Circ_0000423 knockdown suppresses the proliferation, migration and invasion of MGC-803 and AGS cells
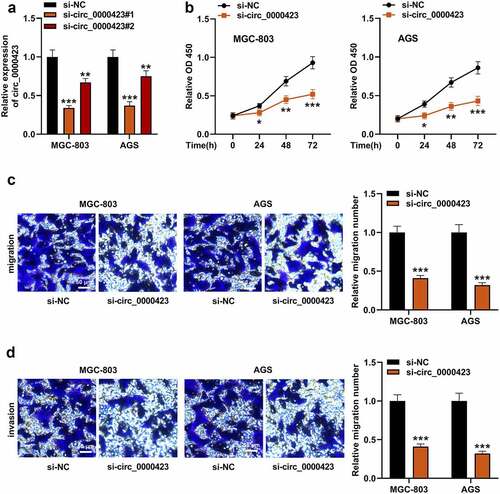
Figure 3. Circ_0000423 directly targets miR-582-3p
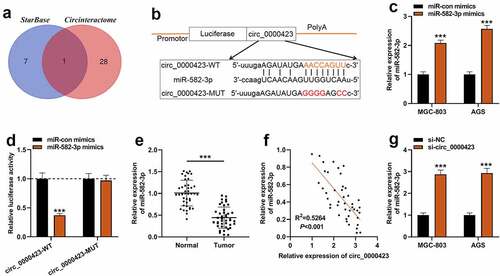
Figure 4. MiR-582-3p inhibition weakens the inhibitory effects of silencing circ_0000423 expression on the malignant biological behaviors of GC cells
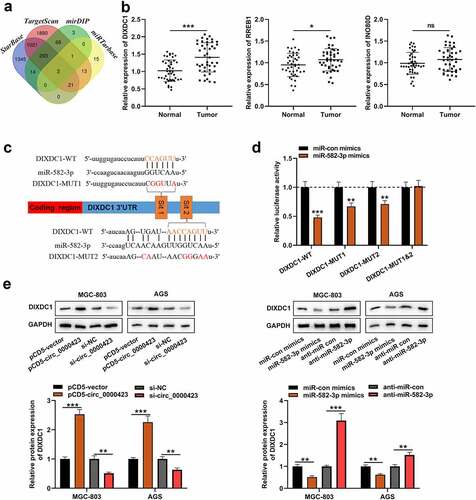
Figure 5. MiR-582-3p targets DIXDC1
Supplemental Material
Download MS Word (1 MB)Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
