Figures & data
Figure 1. Characterization and expression of circMTO1 in oral squamous cell carcinoma (OSCC). (a) The formation of circMTO1 was presented as a diagram. (b) Measurement of circMTO1 and MTO1 expression in transcription inhibitor actinomycin D-treated HOK cells. (c) Measurement of circMTO1 and MTO1 expression in RNase R administrated HOK cells. (d-e) Cellular RNA fractionation (d) and RNA-FISH (e) assays were conducted to assess the cellular distribution of circMTO1. (f) Measurement of circMTO1 expression in OSCC cell lines. (g) Measurement of circMTO1 expression in five cases of OSCC tumor tissues and their adjacent normal tissues. *P < 0.05, **P < 0.01, ***P < 0.001
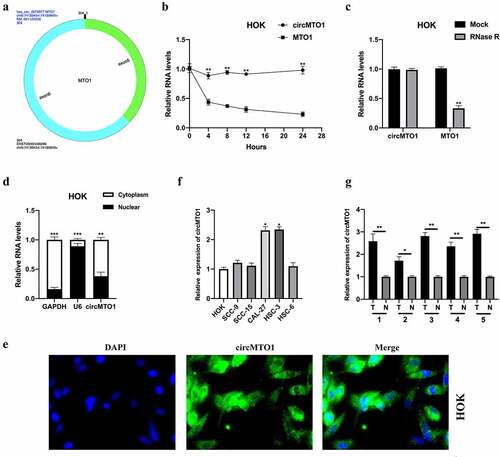
Figure 2. Downregulation of circMTO1 inhibits OSCC tumorigenesis. (a) OSCC cells CAL-27 and HSC-3 were transfected with Sh-NC, Sh-circMTO1#1, and Sh-circMTO1#2 to construct circMTO1 knockdown cell models, and transfection efficiencies were assessed by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR). (b-c) CircMTO1 knockdown CAL-27 and HSC-3 cells were subjected to EDU assay for proliferation ability detection (b), and statistical analysis was conducted (c). (d-e) Transwell migration assay was performed to evaluate circMTO1 knockdown effect on CAL-27 and HSC-3 cells migration level (d), and the results were analyzed (e). (f-g) Transwell invasion experiment was conducted to measure the invasion level of circMTO1 knockdown CAL-27 and HSC-3 cells (f), and the statistical analysis was presented (g). **P < 0.01
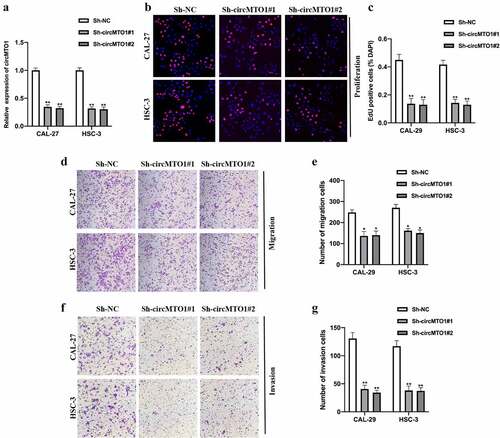
Figure 3. CircMTO1 directly sponges miR-320a. (a) RIP assay using anti-AGO2 and anti-IgG was conducted to assess the miRNA binding possibility of circMTO1, and the results were analyzed by qRT-PCR. (b) Biotinylated RNA pull-down assay was performed to evaluate putative miRNA targets enrichment in circMTO1 probe bounds, and qRT-PCR was used to measure RNAs expression. (c) Wild type (WT) and mutant type (MUT) binding sequences between circMTO1 and miR-320a. (d-e) Dual luciferase reporter assay was applied to assess the interaction between circMTO1 and miR-320a in CAL-27 (d) and HSC-3 (e) cells. (f) Measurement of miR-320a expression in five cases of OSCC tumor tissues and their adjacent normal tissues. (g) Expression of miR-320a in circMTO1 knockdown CAL-27 and HSC-3 cells. *P < 0.05, **P < 0.01, ***P < 0.001
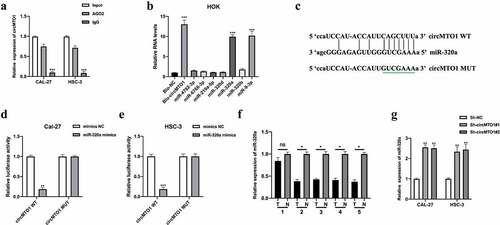
Figure 4. MiR-320a suppresses OSCC tumorigenesis. (a) MiR-320a overexpression cell models were generated by stably transfected NC mimics and miR-320a mimics into CAL-27 and HSC-3 cells, and miR-320a expression was detected by qRT-PCR. (b-c) The proliferation level of miR-320a overexpressed cells were detected by EDU assay (b), and the results were statistically analyzed (c). (d-e) The migration level of miR-320a overexpressed cells were evaluated by transwell assay (d), and the statistical analysis was showed (e). (f-g) The invasion level of miR-320a overexpressed cells was evaluated by transwell assay (f), and the statistical analysis of results was presented (g). *P < 0.05, **P < 0.01, ***P < 0.001
Figure 5. ATRX is a downstream target of miR-320a. The downstream targets of miR-320a were predicted using microT (http://diana.imis.athena-innovation.gr/), PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), and Targetscan (http://www.targetscan.org/mamm_31/) with CLIP Data (strict stringency (≥5)), and degradome Data (high stringency (≥3)). (a) Seven putative mRNA targets were showed. (b) Biotinylated RNA pull-down assay was conducted, and qRT-PCR was used to evaluate putative mRNA targets enrichment in biotinylated miR-320a probe bounds. (c) Predicted binding sites between miR-320a and ATRX. (d-e) The interaction between miR-320a and ATRX was assessed by Dual Luciferase Reporter Assay in HSC-3 (d) and CAL-27 (e) cells. (f-g) Measurement of ATRX mRNA (f) and protein (g) expression in five cases of OSCC tumor tissues and their adjacent normal tissues. (h) Expression level of ATRX in CAL-27 and HSC-3 cells upon miR-320a dysregulation were measured by qRT-PCR. (i-j) Expression level of ATRX in miR-320a dysregulated CAL-27 (G) and HSC-3 (h) cells were detected by western blot. *P < 0.05, **P < 0.01, ***P < 0.001
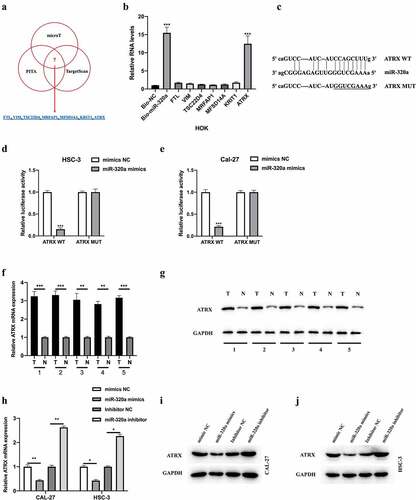
Figure 6. CircMTO1 promotes OSCC progression through miR-320a/ATRX. (a-c) Cell models were constructed by transfecting Sh-NC, Sh-circMTO1#1, Sh-circMTO1#1 + OE-NC, and Sh-circMTO1#1 + OE-ATRX into CAL-27 and HSC-3 cells, and the expression level of ATRX was measured by qRT-PCR (a) and western blot (b and c). (d-e) The proliferation level of treated as indicated cells were measured by EDU assay (d), and statistical analysis results was presented (e). (f-g) The migration level of treated as indicated cells were evaluated by transwell assay (f), and the statistical analysis results is shown (g). (h-i) The invasion level of treated as indicated cells were evaluated by transwell assay (h), and the statistical analysis results was presented (i). *P < 0.05, **P < 0.01, ***P < 0.001
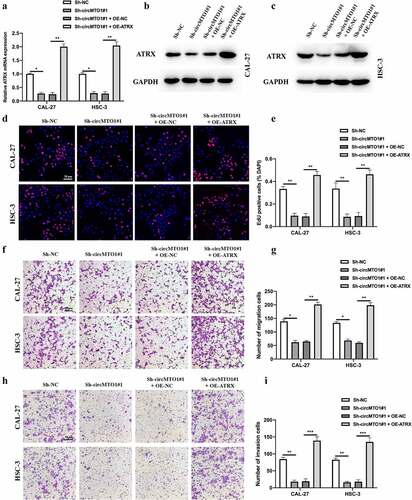
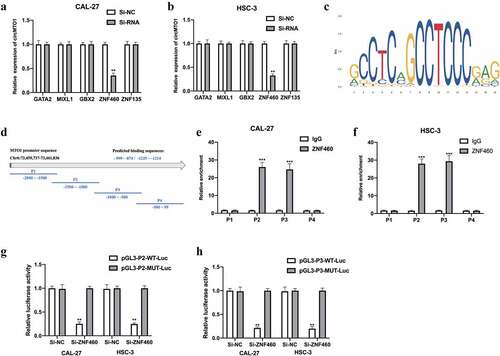
Figure 7. CircMTO1 is transcriptionally upregulated by ZNF460. Transcription factors of circMTO1 were predicted using NCBI (https://www.ncbi.nlm.nih.gov/), UCSC (http://genome.ucsc.edu/), and JASPAR (http://jaspar.genereg.net/) datasets. Five potential factors were predicted (GATA2, MIXL2, GBX2, ZNF460, and ZNF135). (a-b) Expression level of circMTO1 in CAL-27 (a) and HSC-3 (b) cells pre-treated with indicated SiRNAs were measured by qRT-PCR. (c) The predicted binding sites of ZNF460. (d) The promoter of circMTO1 was presented, and the promoter was spliced into P1-P4 section. (e-f) ChIP assay was used in CAL-27 (e) and HSC-3 (f) cells, and the results were analyzed using qRT-PCR. G-(h) CAL-27 (g) and HSC-3 (h) cells were co-transfected with indicated luciferase reporter vectors and siRNAs; luciferase activities were measured. *P < 0.05, **P < 0.01, ***P < 0.001
Availability of data and materials
The datasets and materials used in this study are available based on reasonable requested.
