Figures & data
Table 1. The relationship between SPIN1 expression and clinical features of colorectal cancer patients
Table 2. The sequences of primers for qRT-PCR used in this study
Figure 1. SPIN1 is upregulated in CRC tissues and cell lines. (a) Immunohistochemical staining images of SPIN1 in 90 pairs of paraffin-embedded colorectal cancer (CRC) tissues and adjacent non-cancerous tissues (magnification, 200× and 400×). (b, c) Representative western blotting images and quantitation of SPIN1 protein expression in six CRC cell lines and normal colon cells. (d) The SPIN1 protein expression in 12 pairs of CRC tissues and adjacent normal tissues were assessed by western blotting analysis. (e) The quantification analysis of the blots was analyzed by image J software. P, patient; Student’s t test, *p < 0.05; **p < 0.01.
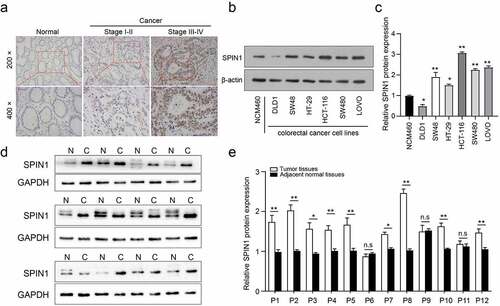
Figure 2. SPIN1 depletion inhibits cell proliferation, migration and invasion. (a) qRT-PCR assays were utilized to confirm the transfection efficiency of siRNAs in HCT-116 and LOVO cells. (b, c) Cell proliferation ability was tested by CCK-8 assays. (d, e) The colony forming assays were used to evaluate the colony formation ability. (f- i) The migration and invasion abilities were evaluated by scratch wound assays (f, g) and transwell invasion assays (h, i) upon transfection of negative control siRNAs and SPIN1 siRNAs, respectively. (j) Representative western blotting analysis of SPIN1 and Wnt/β-catenin pathway components in HCT-116 and LOVO cells upon indicated transfection. Student’s t test, *P < 0.05 and **P < 0.01.
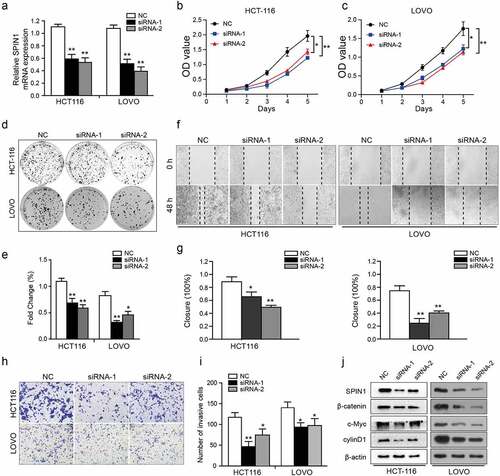
Table 3. The association between miR-381 expression and clinicopathological factors in patients with colorectal cancer
Figure 3. SPIN1 is a target of miR-381 and miR-381 is downregulated in CRC. (a) Construction of the Wild-type (WT) and mutated (MUT) SPIN1 3ʹUTR luciferase reporter vectors. (b, c) The dual-luciferase reporter kit was used to detect the luciferase activity of WT and MUT SPIN1 3ʹUTR plasmids in HCT116 cells and DLD1 cells upon indicated transfection, respectively. (d) The expression of SPIN1 mRNA was assessed by qRT-PCR assays after indicated transfection in CRC cells. (e) Western blotting analysis of SPIN1 protein in CRC cells upon transfection by miR-381 mimics, miR-381inhibitor or negative controls. (f) Relative expression of miR-381 in colorectal cancer (CRC) tissues and paired adjacent tissues (n = 42). Student’s t test, *p < 0.05, **p < 0.01.
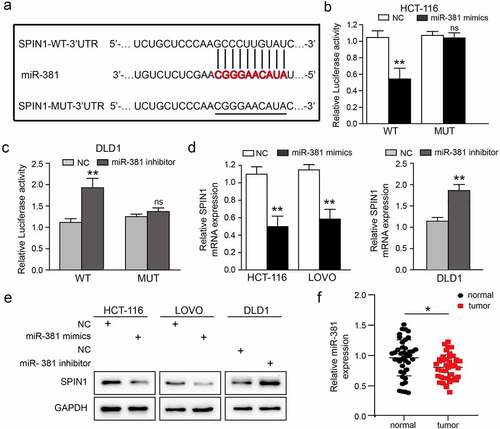
Figure 4. MiR-381 overexpression impedes CRC cell proliferation and invasion. (a) The miR-381 expression in CRC cell lines was evaluated using qRT-PCR assays. (b) Transfection deficiency was validated by qRT-PCR assays in HCT116 and LOVO cells. (c) The well growth abilities of HCT-116 and LOVO were assessed using CCK-8 assays after relative transfection. (d-g) The role of miR-381 on migration and invasion was detected by wound healing and transwell invasion assays. Representative figures (magnification, ×200) and quantification analysis were formed by the indicated groups of cells. (h) Representative western blotting images of SPIN1 and Wnt/β-catenin pathway components following indicated transfection. GAPDH was used as an internal control. Student’s t test, *p < 0.05, **p < 0.01.
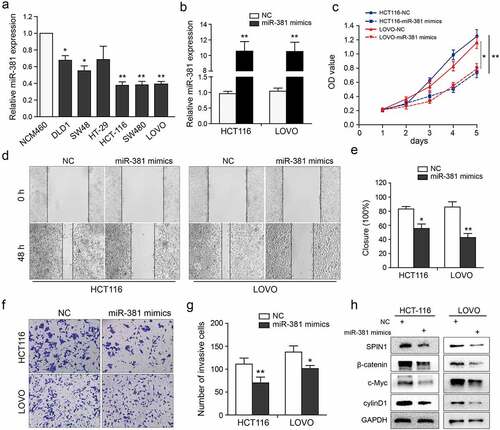
Figure 5. MiR-381 and SPIN1 are responsible for controlling the Wnt/β-catenin signaling pathway in CRC. (a-d) Rescue experiments of overexpressed SPIN1 and miR-381 mimics using with XAV939 or LiCl treatment for 24 h (XAV939,10 μmol/L; LiCl: 20 mmol/L). CCK-8 assays (a), Colony formation assays (b) and transwell invasion assay (c) were conducted to assess cell biological function upon indicated treatments. (d) The protein levels of SPIN1 and Wnt/β-catenin signaling pathway related targets (β-catenin, c-Myc and cyclin D1) were assessed by western blotting upon indicated treatments in LOVO cells. GAPDH was used as an internal control. Student’s t test, *p < 0.05, **p < 0.01.
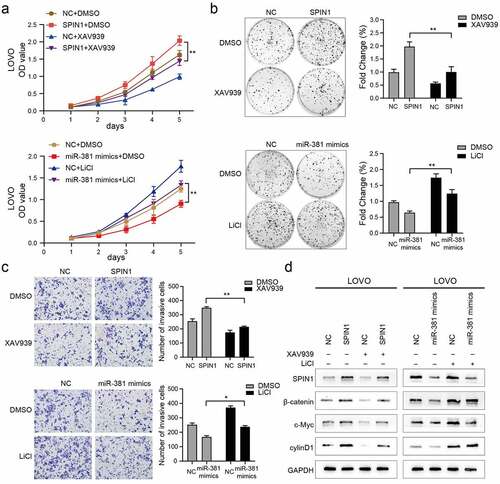
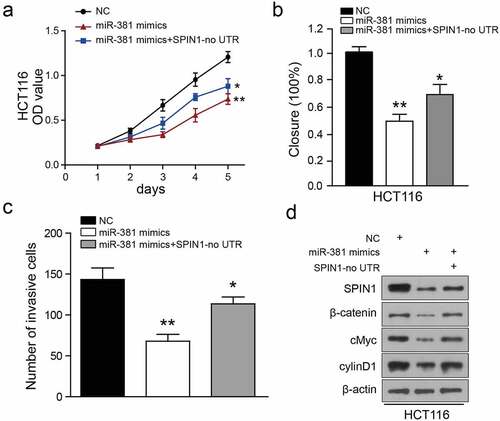
Figure 6. Ectopic expression of SPIN1 attenuates the miR-381-induced effects on colorectal cancer cells. (a) CCK-8 assays performed on miRNA-381-overexpressing HCT-116 cells with SPIN1 overexpression plasmid. (b) Wound healing assays performed on miR-381-overexpressing HCT-116 cells with SPIN1 upregulation. (c) Quantification of the transwell invasion assays under the indicated transfection condition. (d) Representative western blotting images of lysates prepared from miRNA-381-overexpressing HCT-116 cells with SPIN1 upregulation. One-way ANOVA test, *p < 0.05, **p < 0.01.
Availability of data and materials
The datasets used or analyzed during the present study are available from the corresponding authors Ziling Fang and Xiaojun Xiang upon reasonable request.
