Figures & data
Table 1. Expression of a part of DEGs hierarchical clustering analysis results of PCOS group and control group
Figure 1. Summary of DEGs by RNA-seq. (a) X axis represents comparison method between each group. Y axis represents DEG numbers. Red color represents up-regulated DEGs. Blue color represents down-regulated DEGs. (b) Scatter plot of DEGs. X Y axis represents log10 transformed gene expression level, red color represents the up-regulated genes, blue color represents the down-regulated genes, gray color represents the non-DEGs
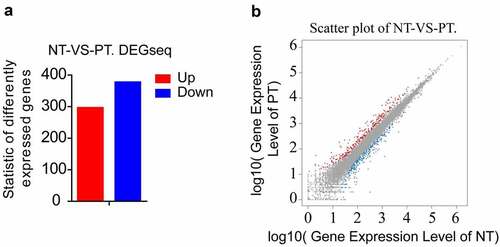
Figure 2. Pathway functional enrichment results of NT-VS-PT. DEGseq Pathway. (a) X axis represents number of DEG. Y axis represents functional classification of KEGG. There are seven branches for KEGG pathways: Cellular Processes, Environmental Information Processing, Genetic Information Processing, Human Disease (For animals only), Metabolism, Organismal Systems and Drug Development. (b) X axis represents enrichment factor. Y axis represents pathway name. The color indicates the q-value (high: white, low: blue), the lower q-value indicates the more significant enrichment. Point size indicates DEG number (The bigger dots refer to larger amount). Rich Factor refers to the value of enrichment factor, which is the quotient of foreground value (the number of DEGs) and background value (total Gene amount). The larger the value, the more significant enrichment
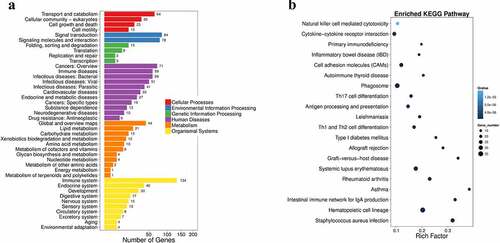
Table 2. Comparison of general conditions between PCOS group and control group
Table 3. Comparison of clinical outcomes between PCOS group and control group
Figure 3. Culture and identification of ovarian GCs in vitro. (a)-(d) represent the situation of GCs cultured in vitro for 1–7 days. (e)-(f) represent the IHC results of GCs staining with FSH antibody during 3–5 days. Scale bar = 100 μm for (e), Scale bar = 50 μm for (f)
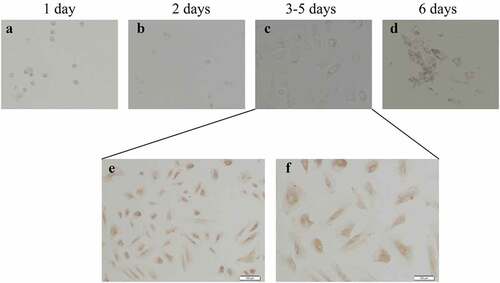
Figure 4. Expression of Hh family members in GCs from PT and control Group. (a)–(d) represent Gli, Ptch1, Gli2 and Gli3 mRNA levels form GCs between normal and PCOS. The red circle represents the normal group (Normal), and the blue circle represents the experimental group (PCOS). p-Values were determined by Student’s t-test, *p < 0.05, **p < 0.01
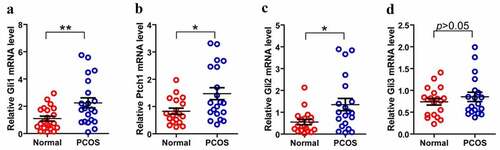
Table 4. Expression of a part of DEGs hierarchical clustering analysis results of PCOS patients and control group
Figure 5. Comparison of TNF-α expression in granulosa cells of two groups. The relative difference of TNF-α mRNA expression results between the normal group (Normal) and the experimental group (PCOS). The red circle represents the normal group (Normal), and the blue circle represents the experimental group (PCOS). * p < 0.05
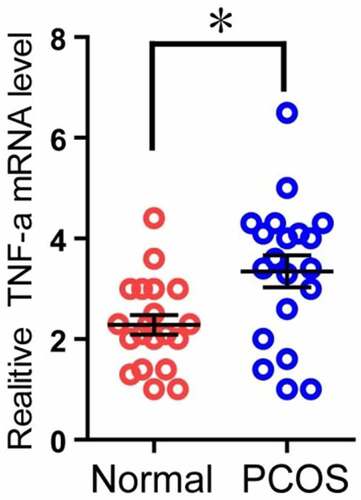
Figure 6. CPA treatment of PCOS ovarian granulosa cells to detect its apoptosis. PCOS cells were treated with KAAD-cyclopamine (CPA, 1um) 24 h later to detect the apoptosis, and then compared with the control group (DMSO). * p < 0.05
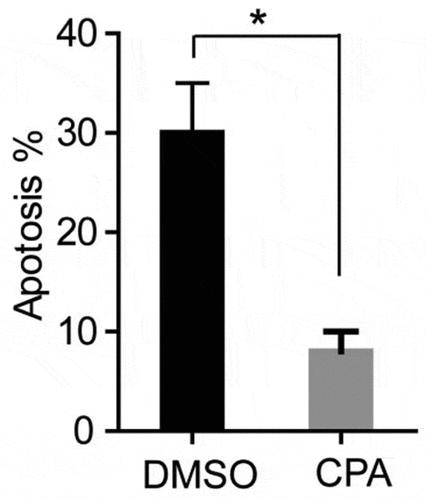
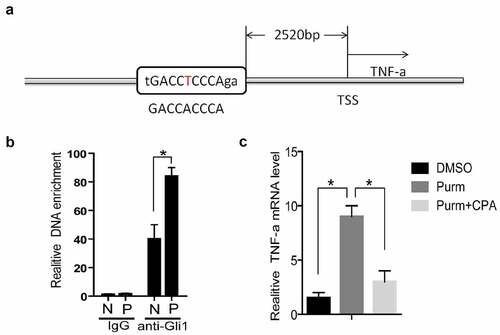
Figure 7. TNF-α is a target gene of Hh signaling pathway. (a) TSS stands for the transcription start site, and its upstream 2520 bases is the Gli binding site predicted by rVista2.0 software, with one base error (red T). (b) In the chromatin immunoprecipitation experiment (ChIP), the Gli1 antibody was used to detect the role of Gli protein in the predicted TNF-α promoter region Gli binding site in the normal group of cells and the experimental group of cells. IgG is the control, anti-Gli1 is immunoprecipitation of Gli1 antibody, N is the normal control group, and P is the experimental PCOS group. *represents p < 0.05. (c) The NIH3T3 cells were treated with DMSO, purm or purm+CPA for 24 hours, and TNF-α mRNA detection was performed. *Represents p < 0.05