Figures & data
Figure 1. Differential gene expression in primary hippocampal neuron inflammation caused by sevoflurane. (a) The flow chart of animal experiment. (b) Inflammatory markers were measured by q-PCR analysis in hippocampus of mice treated by sevoflurane with different concentrations. The results were compared to group 0. (c) Inflammatory markers in mice blood were tested by ELISA assay after treated by sevoflurane with different concentrations. The results were compared to group 0. (d) Differential gene expression in primary hippocampal neuron inflammation. Up-regulated genes with fold change over 1.3 were marked as red dots. Down-regulated genes with fold change less than 0.7 were marked as green dots. (e) 18 top up-regulated genes were validated by q-PCR with or without sevoflurane treatment in mice hippocampus. The results were compared to group 0. (f) 7 top down-regulated genes were validated by q-PCR with or without sevoflurane treatment in mice hippocampus. The results were compared to group 0. (g) Scheme of conservation between Gm5106 and ABCB1. (h) q-PCR analysis of ABCB1 expression with 4% sevoflurane treatment in human SH-SY5Y cell. *p < 0.05, **p < 0.01
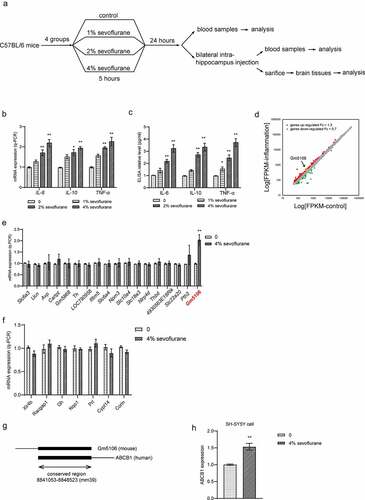
Table 1. The sequence information of siGm5106 (5ʹ-3ʹ)
Table 2. Primers used in q-PCR. (5ʹ to 3ʹ)
Figure 2. Gm5106 related to primary hippocampal neuron inflammation caused by sevoflurane. (a) Gm5106 was silenced by specific siRNA and tested by q-PCR in derived neuron. (b) Cell death rates were measured by Trypan Blue staining after treated with 4% sevoflurane or Gm5106 siRNA in neuron. (c) Inflammatory markers were measured after neuron treated by 4% sevoflurane with Gm5106 siRNA (siGm5106) or a negative control (Neg. Ctr.) in hippocampus of mice. (d) Inflammatory markers in blood samples were measured after mice treated by 4% sevoflurane with Gm5106 siRNA (siGm5106) or a negative control (Neg. Ctr.). **p < 0.01
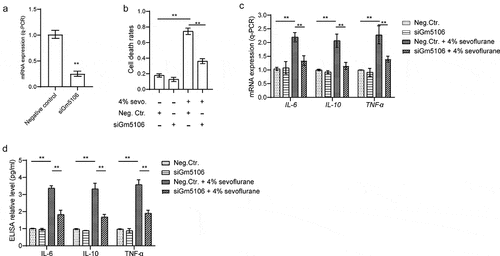
Figure 3. Hoxa5 transcriptionally activated Gm5106. (a) Hoxa5 binding motif was predicted through JASPAR. (b) In HEK-293 T cells, Hoxa5 was overexpressed by pcDNA3.1-Hoxa5 vector and the effect on Gm5106 expression was measured. (c) The effect of sevoflurane on Hoxa5 expression was tested in mice model. (d) The mutation strategy was designed to confirm transcription relations. Two putative binding sites of Hoxa5 in Gm5106 promoter region were mutated and named by M1, M2 and M1/2. (e) Luciferase reporter vectors containing Gm5106 wild type or mutant promoter region were tested. (f) The binding between Hoxa5 and Gm5106 was confirmed by q-ChIP assay, AchR was served as a negative control. (g) The effect of Hoxa5 siRNA on Gm5106 expression with or without 4% sevoflurane treatment was measured. *p < 0.05, **p < 0.01
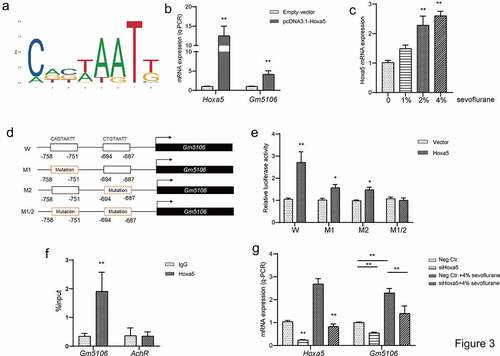
Figure 4. Gm5106 interacted with miR-27b-3p. (a) Gm5106 was predicted as a direct target of miR-27b-3p. (b) The effect of miR-27b-3p mimics on Gm5106 expression was tested by q-PCR. (c) Target between Gm5106 and miR-27b-3p was confirmed by luciferase assay. (d) The effect of sevoflurane on miR-27-3p expression was tested by q-PCR in mice model. (e) The effect of siGm5106 on miR-27b-3p expression in mice with sevoflurane treatment. (f) Inflammatory markers were measured after mice treated by 4% sevoflurane with miR-27b-3p mimics or negative control (Neg. Ctr.). (g) Inflammatory markers in blood samples were measured after neuron treated by 4% sevoflurane with miR-27b-3p mimics or negative control (Neg. Ctr.). *p < 0.05, **p < 0.01
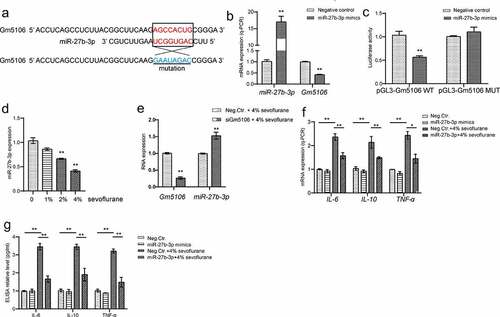
Figure 5. Hoxa5/Gm5106/miR-27-3p positive feedback loop. (a) Hoxa5 was predicted as a direct target of miR-27b-3p. (b) The effect of miR-27b-3p mimics on Hoxa5 expression was tested by q-PCR. (c) The effect of miR-27b-3p mimics on nuclear Hoxa5 protein expression was tested by Western blot. (d) Target between Hoxa5 and miR-27b-3p was confirmed by luciferase assay. (e) Hoxa5 protein and 3ʹUTR overexpression affected Gm5106 and miR-27b-3p expression. (f) Inflammatory markers were measured after neuron treated by 4% sevoflurane with Hoxa5 siRNAs or negative control (scrambled siRNA). (g) Inflammatory markers were measured by ELISA in cell medium after neuron treated by 4% sevoflurane with Hoxa5 siRNAs or negative control (scrambled siRNA). *p < 0.05, **p < 0.01
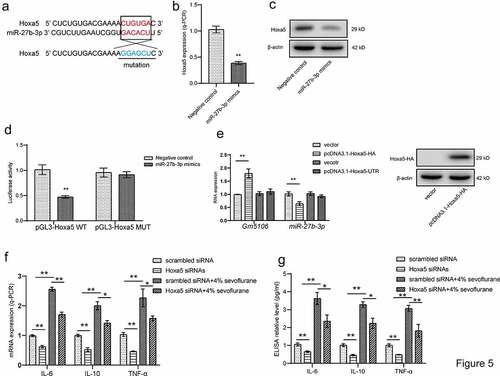
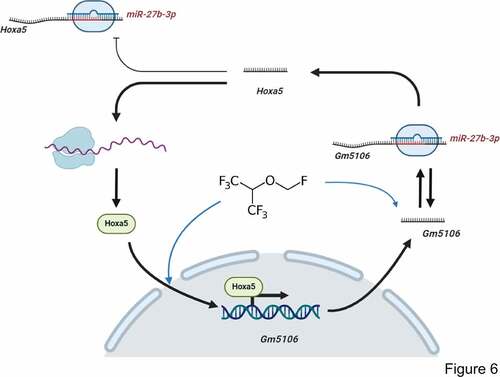
Figure 6. Regulation module of the effect of sevoflurane on neuron inflammation. Sevoflurane induced both long non-coding RNA Gm5106 and TF Hoxa5. During the regulation, Hoxa5 transcriptionally activates Gm5106 which is a direct target of miR-27b-3p. In cytoplasm, Gm5106 interacts with miR-27b-3p and leads to less inhibition of Hoxa5 by miR-27b-3p. Increased Hoxa5 translocates into nucleus to activate Gm5106 and forms positive feedback loop
Data availability
The authors confirm that the data supporting the findings of this study are available within the article.
