Figures & data
Table 1. Relationship between the ITGA11 expression in GC tissue samples and clinical characteristics
Table 2. Primer sequences
Figure 1. Expression of ITGA11 in GC
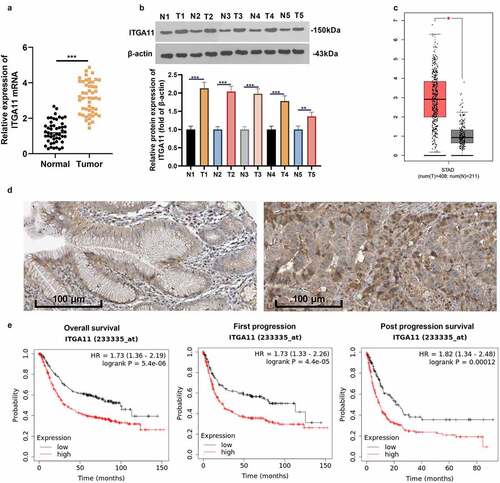
Figure 2. Inhibiting ITGA11 attenuated GC cell proliferation and invasion and strengthened apoptosis
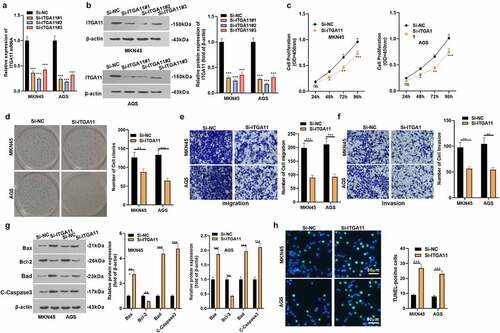
Figure 3. The regulatory effect of ITGA11 on the ITGA11/ PI3K/AKT pathway
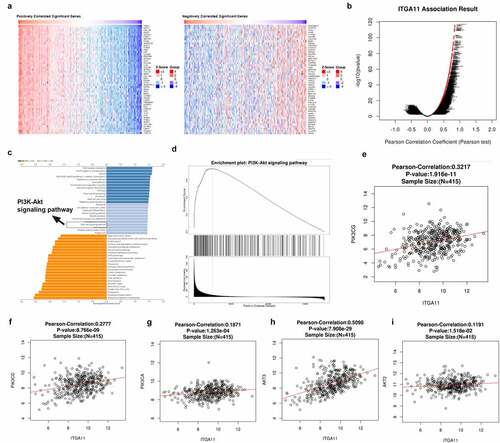
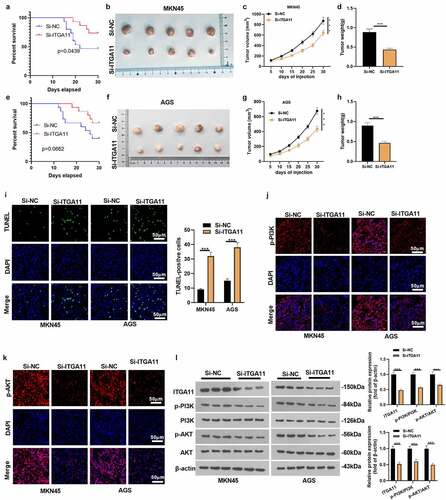
Figure 4. Inhibiting ITGA11 inactivated PI3K/AKT
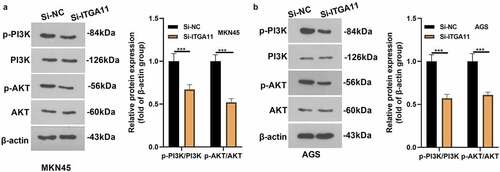
Figure 5. Activating PI3K repressed the effect of ITGA11 knockdown on GC progression
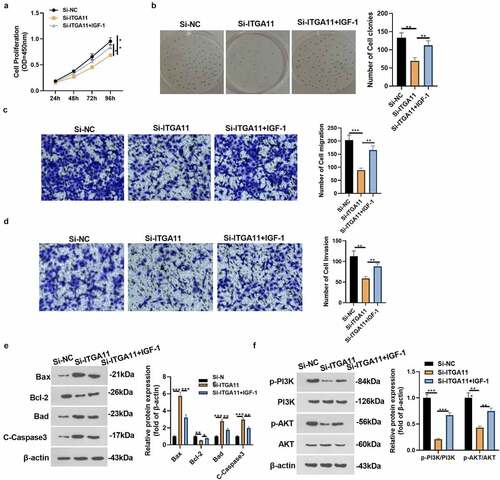
Figure 6. Knocking down ITGA11 dampened GC cell growth cells in vivo.