Figures & data
Table 1. Comparison of the baseline data between the two groups
Figure 1. A total of 105 healthy controls and 102 sepsis patients were included in this study. (a) The expression miR-381-3p in sepsis. (b) The ROC curve showed the diagnosis importance of miR-381-3p. * **P < 0.001
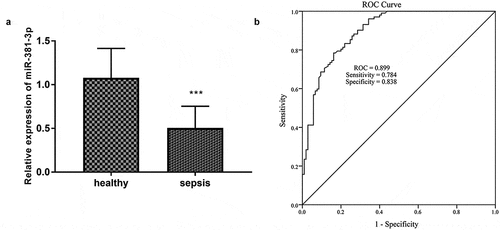
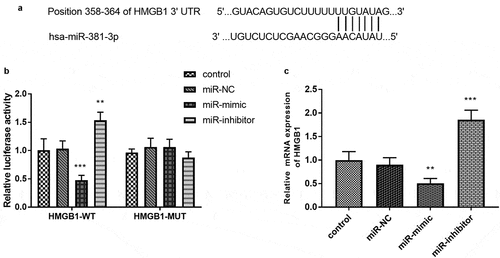
Table 2. Correlation between the levels of miR-381-3p and clinical characteristics in 102 patients with sepsis
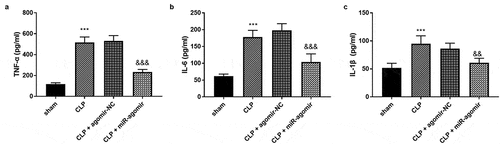
Figure 2. (a) MiR-381-3p mimics reversed the declined expression of miR-381-3p in H9c2 cells. The finding of (b) cell viability, (c) apoptosis, and (d) inflammation in vitro assay. ***P < 0.001, **P < 0.01, compared to control group; &&& P < 0.001, &&P < 0.01, compared to the LPS group
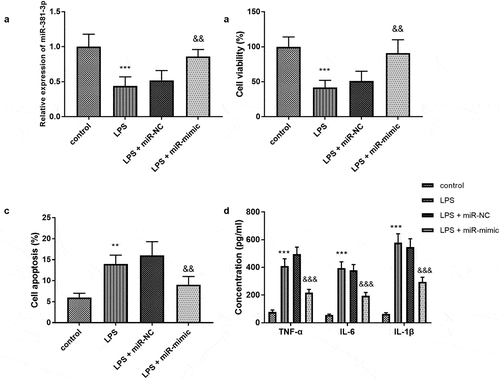
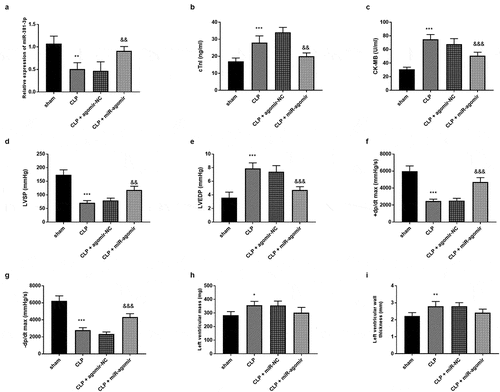
Figure 3. Each group included five rats. (a) MiR-381-3p mimics reversed the declined expression of miR-381-3p in SD rats. The finding of (b) CK-MB, (c) cTnI, (d) LVEDP, (e) LVSP, (f) +dp/dtmax, and (g)-dp/dtmax, (h) LVM, and (i) LVWT. ***P < 0.001, **P < 0.01, *P < 0.05, compared to sham group; &&& P < 0.001, && P < 0.01, compared to the CLP group