Figures & data
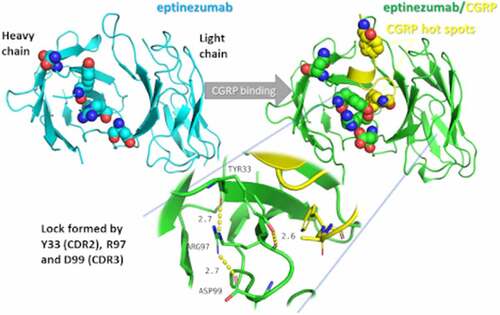
Table 1. X-ray diffraction statistics
Table 2. Model refinement statistics
Table 3. Main epitope/paratope interactions
Figure 1. Crystal structure of eptinezumab in its free (unbound) state, in which the heavy chain (cyan) and the light chain (green) are shown from the side and top views. The CDRs are colored using PyMOL color names, where light chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue])
![Figure 1. Crystal structure of eptinezumab in its free (unbound) state, in which the heavy chain (cyan) and the light chain (green) are shown from the side and top views. The CDRs are colored using PyMOL color names, where light chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue])](/cms/asset/984f3e24-def2-4c94-bccf-d3f30a7e66cb/kbie_a_2006977_f0001_oc.jpg)
Figure 2. Crystal structure of the eptinezumab:CGRP complex. CGRP (yellow) is shown bound in the deep pocket formed by the light (green) and heavy (cyan) chains of the Fab. The CDRs are colored using PyMOL color names, where light chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue])
![Figure 2. Crystal structure of the eptinezumab:CGRP complex. CGRP (yellow) is shown bound in the deep pocket formed by the light (green) and heavy (cyan) chains of the Fab. The CDRs are colored using PyMOL color names, where light chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue])](/cms/asset/acaf2ee1-278e-4df7-a69e-b491cbdf01b0/kbie_a_2006977_f0002_oc.jpg)
Figure 3. Detailed interactions between CGRP and eptinezumab. Hydrogen bond in yellow, hydrophobic contact in pale green, and pi-pi interactions in raspberry. The CDRs are colored such that light chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue]). All colors are PyMOL color names
![Figure 3. Detailed interactions between CGRP and eptinezumab. Hydrogen bond in yellow, hydrophobic contact in pale green, and pi-pi interactions in raspberry. The CDRs are colored such that light chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue]). All colors are PyMOL color names](/cms/asset/27987bc0-e64c-4971-9fa9-32d60ba37470/kbie_a_2006977_f0003_oc.jpg)
Figure 4. Evidence for structural water molecule in eptinezumab:CGRP interface. CGRP is colored in yellow and CDR L2 in Limon, using PyMOL color names. The locations of water molecules from the X-ray crystallography structure of the uncomplexed eptinezumab (orange spheres), of water molecules obtained by Watermap from the complex structure (light blue spheres 1–3), and of explicit clusters of water molecules obtained by Watermap from the eptinezumab:CGRP complex (HOH clusters). Watermap places 2 water molecules (light blue spheres 1 and 2) in a cavity within the bound structure, forming a network of hydrogen bond between Asp51 NH (CDR L2) to water1, water1 to water2, water2 to Leu90 C = O and Ser92 OG (CDR L3), and finally Ser92 OGH to Ser116 C = O (CGRP); CGRP Ser116 OGH does not make any direct interaction with eptinezumab but faces another small cavity. Watermap placed one water molecule (light blue sphere 3) in this cavity, indicating a hydrogen bond network with O from water3 and water3 to Cys100 and Tyr93 (CDR L3) carbonyls
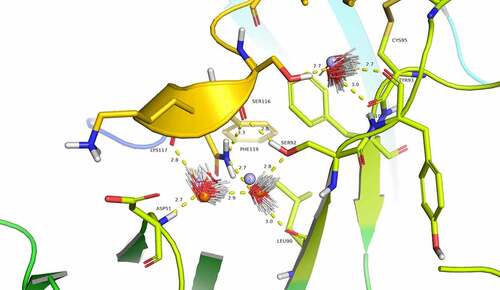
Figure 5. Comparison of bound and unbound structures indicate conformational changes during eptinezumab binding to CGRP. Eptinezumab is in yellow. The CDRs are colored using PyMOL color names, where light-chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy-chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue]). Superposed structure using the FV domain as template a) of the unbound (translucent) and bound (opaque) structure. CDR H3 (blue) clearly goes through a structural reorganization where an internal H-bond between Asp99 and Arg97 in the bound form goes to an H-bond between Asp99 and light chain Lys46 inducing a movement of Asp99 CA by ~3 Å (from 2.5 Å to 3.5 Å following the alignment); b) of the unbound (translucent) and bound (opaque). Asn54 rotates and makes a direct contact with CGRP Asn113. Although the angles between both amides is not right for a proper hydrogen bond, the inter distance between N-Asn113 and O from Asn54 CDR H2 is 2.8 Å
![Figure 5. Comparison of bound and unbound structures indicate conformational changes during eptinezumab binding to CGRP. Eptinezumab is in yellow. The CDRs are colored using PyMOL color names, where light-chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy-chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue]). Superposed structure using the FV domain as template a) of the unbound (translucent) and bound (opaque) structure. CDR H3 (blue) clearly goes through a structural reorganization where an internal H-bond between Asp99 and Arg97 in the bound form goes to an H-bond between Asp99 and light chain Lys46 inducing a movement of Asp99 CA by ~3 Å (from 2.5 Å to 3.5 Å following the alignment); b) of the unbound (translucent) and bound (opaque). Asn54 rotates and makes a direct contact with CGRP Asn113. Although the angles between both amides is not right for a proper hydrogen bond, the inter distance between N-Asn113 and O from Asn54 CDR H2 is 2.8 Å](/cms/asset/f9a2421a-c79b-4082-954c-c9c867fceda8/kbie_a_2006977_f0005_oc.jpg)
Figure 6. Computational alanine scanning of CGRP identifies key interface residues. a) The CDRs are colored using PyMOL color names, where light-chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy-chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue]). CGRP is in gray. Four positions in CGRP are important for binding: Phe109 (red), Val114 (Orange), Gly115 (yellow), Phe119 (green). b) Table showing loss in binding energetics observed when these key positions mutated to the corresponding amino acid in related neuropeptides based on sequence alignment
![Figure 6. Computational alanine scanning of CGRP identifies key interface residues. a) The CDRs are colored using PyMOL color names, where light-chain regions are in shades of green (L1 [pale green], L2 [lemon], L3 [lime]) and heavy-chain regions are in shades of blue (H1 [light blue], H2 [marine], H3 [blue]). CGRP is in gray. Four positions in CGRP are important for binding: Phe109 (red), Val114 (Orange), Gly115 (yellow), Phe119 (green). b) Table showing loss in binding energetics observed when these key positions mutated to the corresponding amino acid in related neuropeptides based on sequence alignment](/cms/asset/de5325f7-aafb-4701-a5e2-5623e012842d/kbie_a_2006977_f0006_oc.jpg)
