Figures & data
Table 1. Clinical features of 30 glioma patients and the expression of IP6K2
Figure 1. IP6K2 exhibited high expression in glioma tissues A Analyze the expression level of IP6K2 in a variety of tumors through the TIMER2.0 database; B Analyze the expression of IP6K2 in 163 glioma tissues and 207 normal tissues through the TCGA database; C Analyze the expression of IP6K2 in 23 glial tissues through the Oncomine database The expression of tumor tissue and 81 normal tissues; D Analyze the correlation between IP6K2 expression and the overall survival rate (OS) of glioma patients through TCGA database; E Analyze the relationship between IP6K2 expression level and disease-free survival of glioma patients through TCGA database (DFS) correlation; F Detected the mRNA expression of IP6K2 in glioma tissues and normal control tissues by RT-PCR assay; G Detected the protein expression of IP6K2 in glioma tissues and normal control tissues by Western blot assay. *p < 0.05; ** p < 0.01; *** p < 0.001.
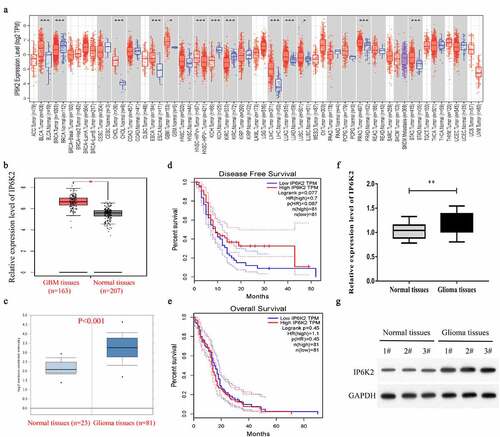
Figure 2. Overexpression of IP6K2 could promote cells to proliferate, migrate and invade A The mRNA expression of IP6K2 in glioma cells and normal control cells was detected by RT-PCR; B U87 and U251 were down-regulated by small interfering RNA (sh-IP6K2-1, sh-IP6K2-2 and sh-IP6K2-3) The expression of IP6K2 in the cells was verified by RT-PCR; C The expression of IP6K2 in U87 and U251 cells was up-regulated by overexpression plasmids and verified by RT-PCR; D Staining efficiency was verified by Western blot experiments for the conversion of sh-IP6K2-1 and OE-IP6K2; E The effect of IP6K2 on the proliferation of U87 and U251 cells was tested by EdU experiment. (Magnification: 200X) F-G Transwell experiment was conducted to detect changes in cell migration and invasion ability after inhibiting or overexpressing IP6K2 in U87 and U251 cell lines. (Magnification: 200X) *p < 0.05; ** p < 0.01.
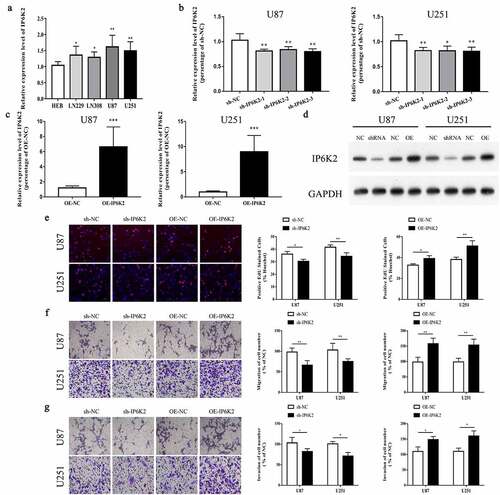
Figure 3. Overexpression of IP6K2 promoted the growth of glioma in vivo A Representative images of tumors in nude mice in each group; B Determine tumor volume; C Determine tumor weight; D RT-PCR showed that the expression of IP6K2 was increased in the tumor tissues of the LV-IP6K2 group; E Western blot showed that IP6K2 was expressed in the LV-IP6K2 group Increased in the tumor tissue. *p < 0.05.
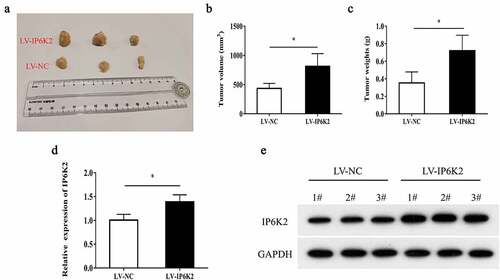
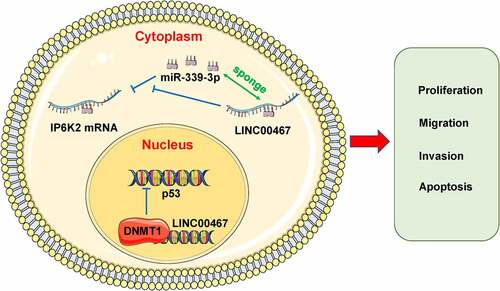
Figure 4. MiR-339-3p could bind to LINC00467 and IP6K2 at the same time A Bioinformatics predicts miRNAs that may bind to LINC00467 and IP6K2, including miR-2467-3p, miR-339-3p, miR-1251-5p, miR-378 and miR-217; B Discover miR on the Kean side of the RIP experiment −339-3p has the strongest binding ability with LINC00467; C Construct LINC00467 wild-type plasmid (LINC00467-WT) and LINC00467 mutant plasmid (LINC00467-MUT) according to the binding site; D It is found through the double luciferase reporter gene experiment test, miR-339-3p can be combined with LINC00467; E Construction of IP6K2 wild-type plasmid (IP6K2-WT) and IP6K2 mutant plasmid (IP6K2-MUT) based on the binding site; D Through the double luciferase reporter gene test, it was found that miR- 339-3p can bind to IP6K2. *p < 0.05; **p < 0.01.
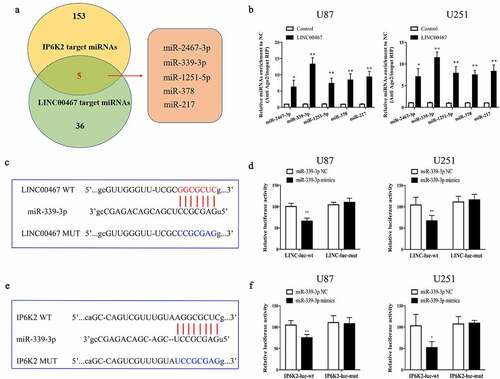
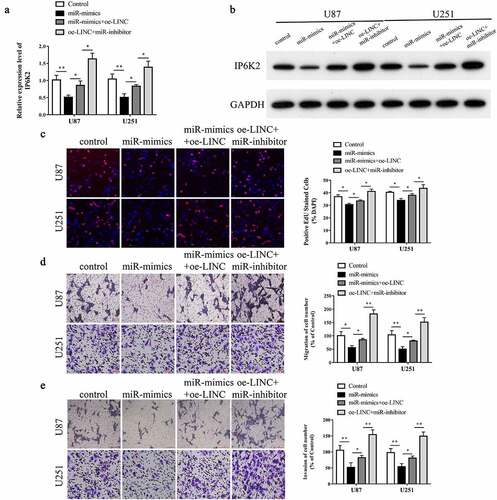
Figure 5. LINC00467/miR-339-3p/IP6K2 regulatory axis promoted the proliferation, migration and invasion of glioma cells A-B After U87 and U251 cells were simultaneously transfected with miR-339-3p mimics, miR-339-3p inhibitor and LINC00467 overexpression plasmids, the expression of IP6K2 was detected by RT-PCR and Western blot experiments; C The effect of LINC00467/miR −339-3p/IP6K2 regulation axis on the proliferation of glioma cells was detected by EdU experiment; D-E Transwell experiment was used to detect the effect of LINC00467/miR-339-3p/IP6K2 regulation axis on the migration and invasion of glioma cells. *p < 0.05; ** p < 0.01.