Figures & data
Table 1. Sequences of primers of qPCR, si-RNA and miRNA
Figure 1. Expression of LINC00525 in colorectal cancer cells.
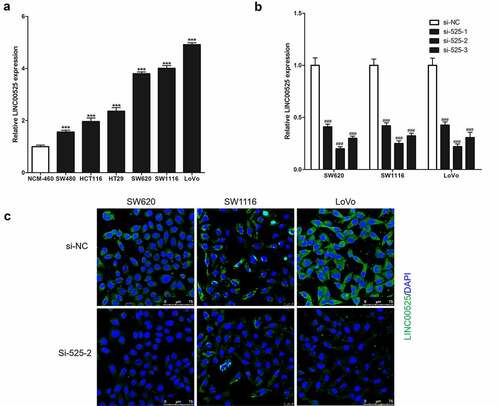
Figure 2. LINC00525 promoted the proliferation of colorectal cancer cells in vitro and in vivo.
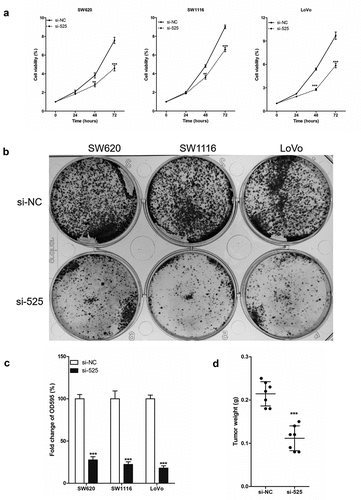
Figure 3. LINC00525 promoted hypoxic glycolysis in colorectal cancer cells.
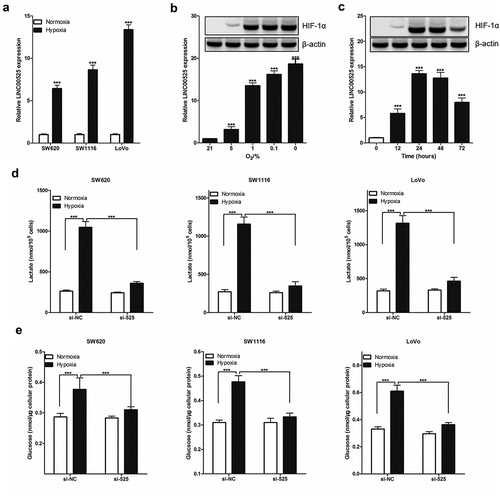
Figure 4. Effect of LINC00525 on hypoxic glycolysis is HIF-1α dependent.
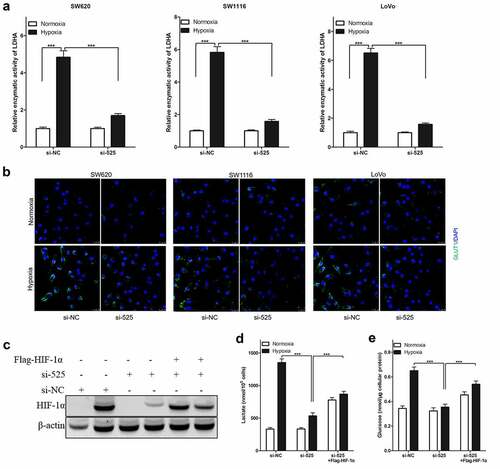
Figure 5. LINC00525 promoted UBE2Q1 expression by sponging to miR-338-3p.
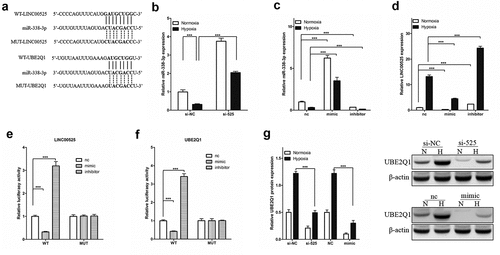
Figure 6. Effect of LINC00525 on hypoxic glycolysis is UBE2Q1 dependent.
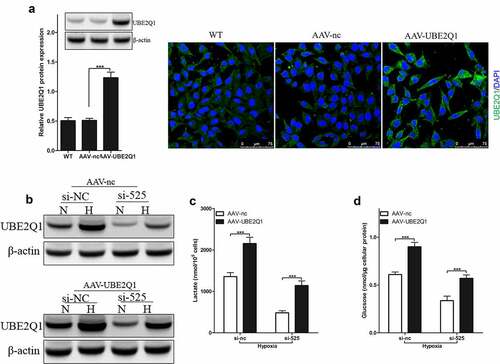
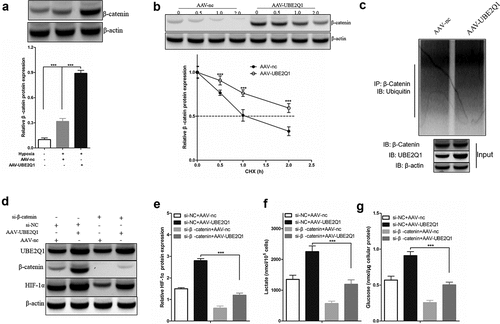
Figure 7. UBE2Q1 stabilized β-catenin, and β-catenin knockdown hypoxia-induced abrogated LINC00525-augmented HIF-1α activation and glycolysis.