Figures & data
Table 1. Number of DE mRNA, tRF/tiRNA and miRNA in PAITA
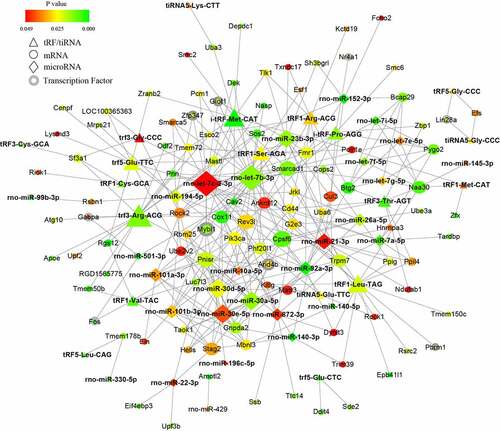
Figure 2. The microRNA-mRNA regulatory network in PAITA. Triangle represents microRNA. Circle represents mRNA. Circle with gray edge represents transcription factor. Color represents the p value of DE genes. Node size represents network degree of DE gene.
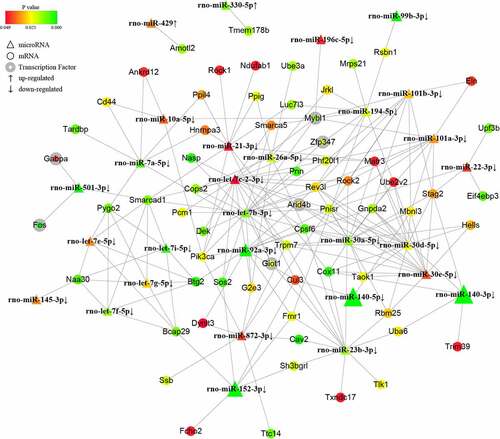
Figure 3. The tRF/tiRNA-mRNA regulatory network in PAITA. Triangle represents tRF/tiRNA. Circle represents mRNA. Circle with gray edge represents transcription factor. Color represents the p value of DE genes. Node size represents network degree of DE gene.
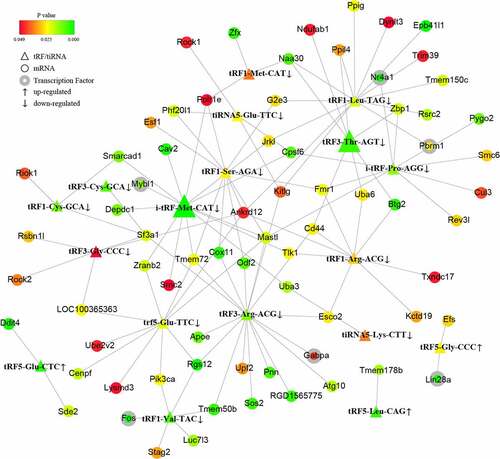
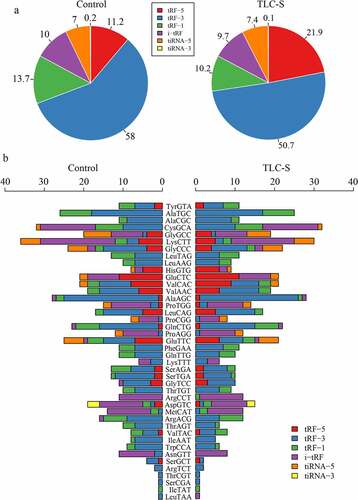
Figure 7. The distribution of six tRF/tiRNA types in PAITA. a, Overall distribution of six tRF/tiRNA types. b, Distribution of six tRF/tiRNA types in each tRNA.