Figures & data
Table 1. Primer sequence
Table 2. Association of miR-545-3p and clinicopathological features of CRC patients
Figure 1. MiR-545-3p is silenced in CRC. (A) Test of the relativity of miR-545-3p in paired CRC and adjacent normal tissues was via reverse transcription quantitative PCR (n = 67). (B) Examination of miR-545-3p in CRC cell lines and NCM460 cells was via qPCR; (C) Kaplan Meier survival analysis manifested elevated miR-545-3p was linked with poor prognosis in CRC patients. * P< 0.05 versus the NCM460 cells. The data in the figures were all measurement data in the form of mean ± SD.
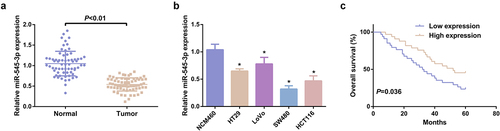
Figure 2. SHK suppresses the progression with facilitated autophagy of CRC cells. (A) Determination of the influence of SHK on cell viability was via MTT method. (B) Treatment of the cells was with different concentrations of SHK for 24 h, and test of the cell cycle was via flow cytometry. (C, D) Examination of cell migration and invasion was via Transwell. (E) Detection of cell apoptosis was via Flow cytometry. (F) Analysis of autophagy-correlated protein was via Western blot, GAPDH presented as load control. Treatment of SW480 cells was with diverse concentrations of SHK medium. N = 3; * P < 0.05 versus the 0 mg/mL concentration of SHK. The data in the figures were all measurement data in the form of mean ± SD.
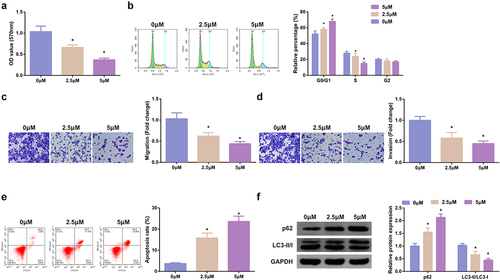
Figure 3. MiR-545-3p inhibitor turns around the action of SHK on CRC cells. (A) CRC cells were introduced with different concentrations of SHK medium to detect miR-545-3p in the cells. (B) Detecting miR-545-3p transfection efficiency via qPCR. (C) Determination of the impact of repressing miR-545-3p on cell viability was via MTT method. (D) Examination of the cell cycle variation after suppression of miR-545-3p was via Flow cytometry. (E, F) Test of cell migration and invasion after repressed miR-545-3p was via Transwell. (G) Examination of cell apoptosis after repression of miR-545-3p via Flow cytometry. (H) Analysis of autophagy-correlated protein was via Western blot, GAPDH presented as load control. N = 3; * P < 0.05 versus the 0 mg/ml concentration of SHK; + P< 0.05 versus the in-NC. The data in the figures were all measurement data in the form of mean ± SD.
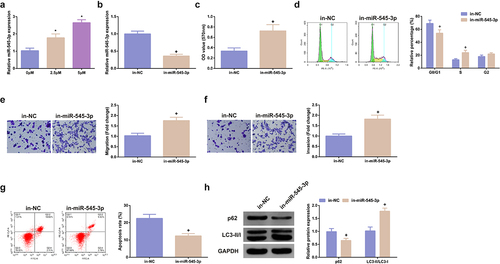
Figure 4. MiR-545-3p targets GNB1. (A) Sequence alignment was of miR-545-3p with GNB1 3ʹUTR-WT and GNB1 3ʹUTR-MUT. (BC) GNB1 mRNA in CRC and cell lines. (D) Pearson correlation analysis elucidated reverse association of miR-545-3p with GNB1 mRNA in the collected tumor tissue samples. (EF) GNB1 mRNA and protein in cells was elevated behind transfection of miR-545-3p inhibitor. (G) Elevated miR-545-3p in cells repressed the relative activity of GNB1 3ʹUTR-WT luciferase. (H) Test of CRC cells was via RNA immunoprecipitation with AGO2 antibody. The results of reverse transcription quantitative PCR clarified that AGO2 simultaneously enriched GNB1 mRNA and miR-545-3p. N = 3; * P < 0.05 versus the NCM460 cells; + P< 0.05 versus the in-NC. The data in the figures were all measurement data in the form of mean ± SD.
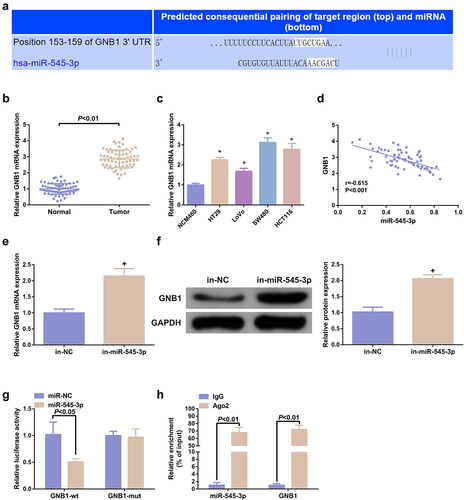
Figure 5. Elevation of GNB1 turns around the tumor suppressor effect of SHK on CRC cells. (A) Examination of the transfection efficiency of elevating GNB1 was via qPCR. (B) Determination of the impact of elevation of GNB1 on cell viability was via MTT method. (C) Detection of cell cycle variation after augmentation of GNB1 was via Flow cytometry. (D, E) Detection of the migration and invasion of cells after elevation of GNB1 was via Transwell. (F) Test of cell apoptosis after elevation of GNB1 was via Flow cytometry. (G) Analysis of autophagy-related protein was via Western blot, GAPDH presented as load control. N = 3; # P< 0.05 versus the oe-NC; & P < 0.05 versus the in-miR-545-3p + si-NC. The data in the figures were all measurement data in the form of mean ± SD.
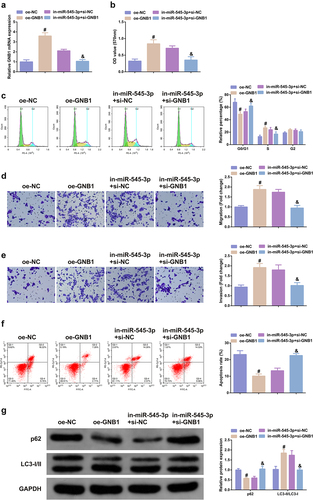
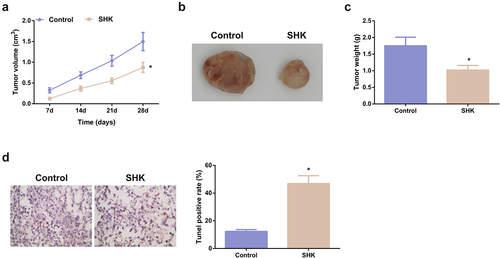
Figure 6. SHK represses tumor advancement. (A, B) The volume and tumor map of xenogeneic tumors. (C) The weight of the xenogeneic tumor. (D) Examination of tumor tissue apoptosis was with TUNEL staining. N = 6; *P < 0.05 versus the control. The data in the figures were all measurement data in the form of mean ± SD.