Figures & data
Table 1. The characteristics of the endometrial carcinoma patients
Figure 2. Hsa_circ_0039569 was elevated in endometrial carcinoma. (a) Expression of hsa_circ_0039569 in normal tissues and endometrial carcinoma tissues was detected by qRT–PCR. (b) Illustration of the generation of hsa_circ_0039569 from the CCL22 gene. (c) Expression of hsa_circ_0039569 in endometrial carcinoma cell lines was detected by qRT–PCR. (d) The distribution of hsa_circ_0039569 in the nucleus and cytoplasm was detected by qRT–PCR. * denoted P < 0.05, ** denoted P < 0.01, *** denoted P < 0.001.
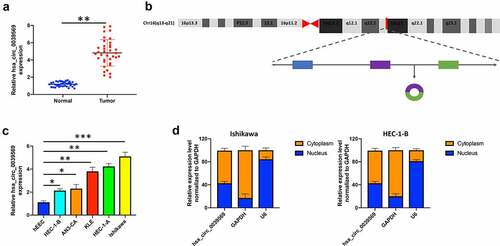
Figure 3. Hsa_circ_0039569 regulated cell proliferation in endometrial carcinoma. Hsa_circ_0039569 was overexpressed in HEC-1-B cells and was knocked down in Ishikawa cells. (a) Expression of hsa_circ_0039569 in HEC-1-B and Ishikawa cells was detected by qRT–PCR. (b) Cell viability was determined by CCK8. (c) A colony formation assay was performed to determine cell proliferation. (d) Cell apoptosis was evaluated by flow cytometry. * denoted P < 0.05, ** denoted P < 0.01.
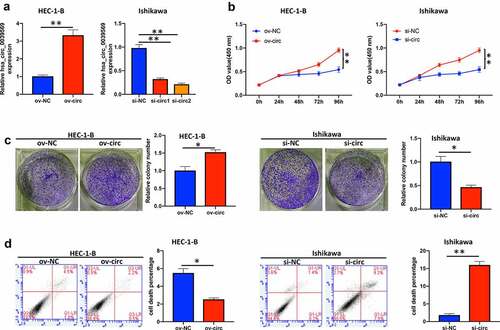
Figure 4. Hsa_circ_0039569 mediated the migration and invasion of endometrial carcinoma cells. Hsa_circ_0039569 was overexpressed in HEC-1-B cells and knocked down in Ishikawa cells. (a and b) Cell migratory ability was assessed by wound healing (a) and Transwell (b) assays. (c) Cell invasive ability was assessed by Transwell assay. * denoted P < 0.05.
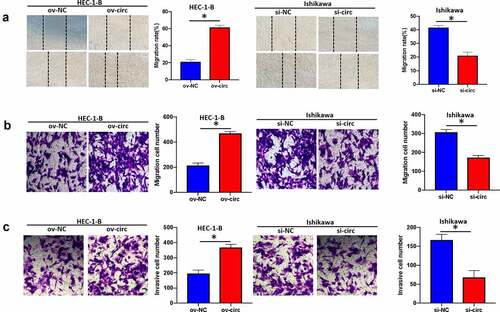
Figure 5. Hsa_circ_0039569 acts as a sponge of miR-197. (a) Expression of miR-197 in normal tissues and endometrial carcinoma tissues was detected by qRT–PCR. (b) The correlation between miR-197 and hsa_circ_0039569 was assessed by Pearson’s correlation coefficient test. (c) MiR-197 was silenced in HEC-1-B cells and overexpressed in Ishikawa cells. (d) Illustration of the predictive binding site between miR-197 and hsa_circ_0039569. (e) RIP assay was performed to validate the direct binding of miR-197 and hsa_circ_0039569 on Ago2. (f) A dual luciferase reporter assay was performed to verify the interaction of miR-197 and hsa_circ_0039569. (g) Hsa_circ_0039569 was overexpressed in HEC-1-B cells and knocked down in Ishikawa cells. Expression of miR-197 was detected by qRT–PCR. * denoted P < 0.05, ** denoted P < 0.01.
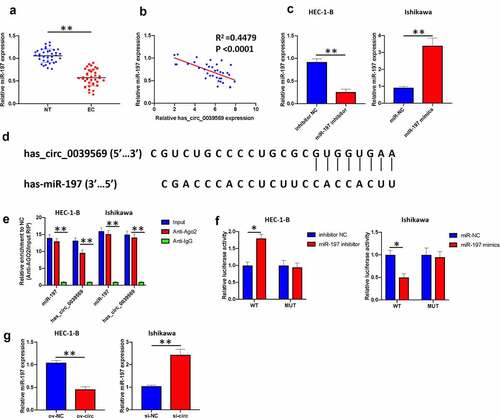
Figure 6. Hsa_circ_0039569 regulated the expression of HMGA1 by targeting miR-197. (a) Expression of HMGA1 in normal tissues and endometrial carcinoma tissues was detected by qRT–PCR. (b) Correlations between miR-197 and HMGA1 or HMGA1 and hsa_circ_0039569 were assessed by Pearson’s correlation coefficient test. (c) Illustration of the predictive binding site between miR-197 and HMGA1. (d) A dual luciferase reporter assay was performed to verify the interaction of miR-197 and HMGA1. (e and f) Hsa_circ_0039569 was overexpressed in HEC-1-B cells with miR-197 mimics or was knocked down in Ishikawa cells with a miR-197 inhibitor. The level of HMGA1 was evaluated by qRT–PCR (e) and Western blot (f). * denoted P < 0.05, ** denoted P < 0.01.
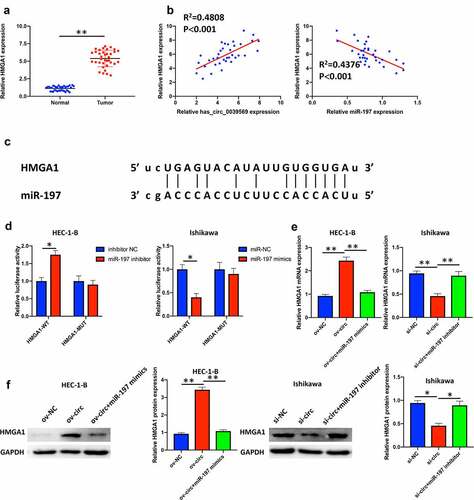
Figure 7. Hsa_circ_0039569 regulated cell proliferation in endometrial carcinoma via the miR-197/HMGA1 axis. (a and b) HMGA1 was knocked down by siRNA in HEC-1-B cells or overexpressed in Ishikawa cells. The level of HMGA1 was evaluated by qRT–PCR (a) and Western blot (b). (C to E) Hsa_circ_0039569 was overexpressed in HEC-1-B cells with miR-197 mimics or HMGA1 knockdown. Hsa_circ_0039569 was knocked down in Ishikawa cells overexpressing miR-197 inhibitor or HMGA1. (c) Cell viability was determined by CCK8. (d) A colony formation assay was performed to determine cell proliferation. (e) Cell apoptosis was evaluated by flow cytometry. * denoted P < 0.05, ** denoted P < 0.01.
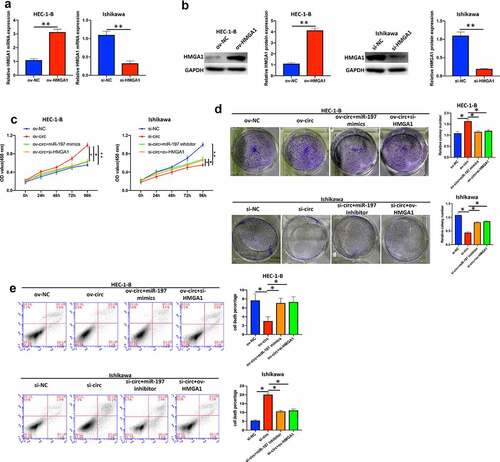
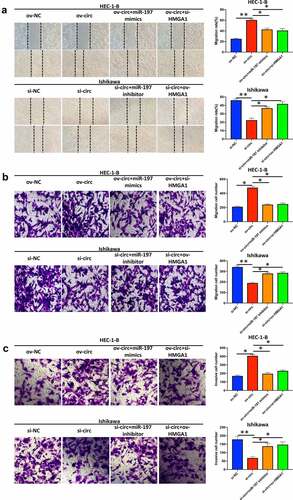
Figure 8. Hsa_circ_0039569 regulated the migration and invasion of endometrial carcinoma cells via the miR-197/HMGA1 axis. Hsa_circ_0039569 was overexpressed in HEC-1-B cells with miR-197 mimics or HMGA1 knockdown. Hsa_circ_0039569 was knocked down in Ishikawa cells overexpressing miR-197 inhibitor or HMGA1. (a and b) Cell migrative ability was assessed by wound healing (a) and Transwell (b) assays. (c) Cell invasive ability was assessed by Transwell assay. * denoted P < 0.05, ** denoted P < 0.01.
Data availability statement
The datasets used or analyzed in the current study are available from the corresponding author on reasonable request.