Figures & data
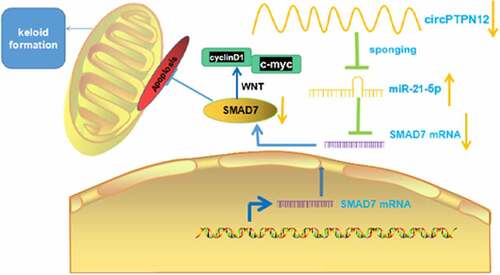
Figure 1. CircPTPN12 declined in keloid tissue and keloid fibroblasts, and silencing circPTPN12 could promote the growth of keloid fibroblasts. (a) qRT-PCR was applied to detect the expression of circPTPN12 in keloid tissue. (b) qRT-PCR was applied to detect the expression of circPTPN12 in keloid fibroblasts. (c) qRT-PCR was used to detect the transfection efficiency of circPTPN12 knockdown. (d) MTT assay was applied to assess the cell viability of keloid fibroblasts. (e) Flow cytometry was used to detect cell apoptosis of keloid fibroblasts. (f) Transwell assay was applied to assess the cell migration and invasion of keloid fibroblasts. ** P < 0.01, *** P < 0.001.
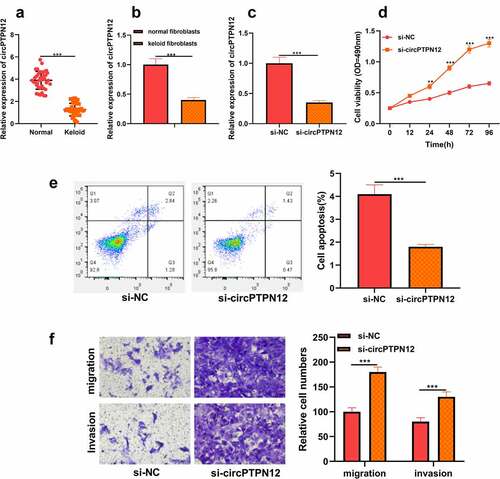
Figure 2. CircPTPN12 could sponge miR-21-5p in keloid fibroblasts. (a) CircPTPN12 contained a conserved binding site of miR-21-5p. (b) qRT-PCR was applied to detect the expression of miR-21-5p in keloid tissue. (c) qRT-PCR was applied to detect the expression of miR-21-5p in keloid fibroblasts. (d) The binding of circPTPN12 and miR-21-5p was verified by dual-luciferase reporter gene assay. (e, f)The combination of circPTPN12 and miR-21-5p was verified by RNA immunoprecipitation and pull-down assays. (g) qRT-PCR was used to detect the level of miR-21-5p after low expression of circPTPN12. (h) Pearson correlation analysis was used to detect the correlation between circPTPN12 and miR-21-5p in keloid tissue. *** P < 0.001.
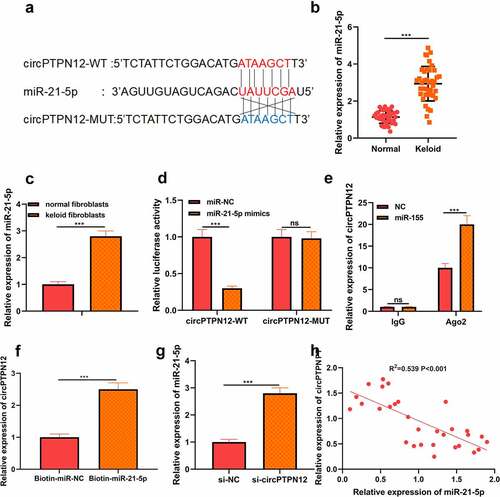
Figure 3. MiR-21-5p directly targeted SMAD7 in keloid fibroblasts. (a) SMAD7 contained a conserved binding site of miR-21-5p. (b) The binding of SMAD7 and miR-21-5p was verified by dual-luciferase reporter gene assay. (c) qRT-PCR was applied to detect the expression of SMAD7 in keloid tissue. (d) qRT-PCR was applied to detect the expression of SMAD7 in keloid fibroblasts. (e) Pearson correlation analysis was used to detect the correlation between SMAD7 and miR-21-5p in keloid tissue. (f) qRT-PCR was used to detect the level of SMAD7 after overexpression of miR-21-5p. Cotransfection of miR-21-5p inhibitors and si-SMAD7 into keloid fibroblasts. (g) The levels of miR-21-5p and SMAD7 were detected by qRT-PCR. (h) MTT assay was applied to assess the cell viability of keloid fibroblasts. (i) Flow cytometry was used to detect cell apoptosis of keloid fibroblasts. (j) Transwell assay was applied to assess the cell migration and invasion of keloid fibroblasts. * P < 0.05, ** P < 0.01, *** P < 0.001.
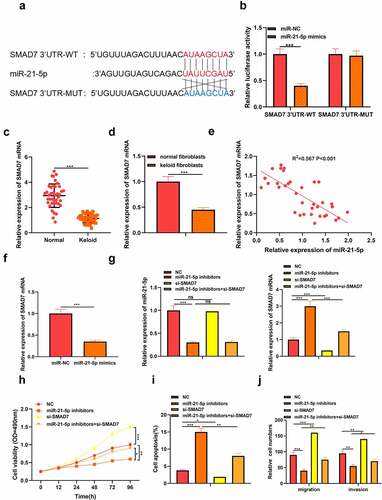
Figure 4. Silencing circPTPN12 targeted miR-21-5p/SMAD7 axis and activated the Wnt pathway to strengthen keloid fibroblasts’ growth. Cotransfection of miR-21-5p inhibitors and si-circPTPN12 into keloid fibroblasts. (a) The level of SMAD7 was detected by qRT-PCR. (b) MTT assay was applied to assess the cell viability of keloid fibroblasts. (c) Flow cytometry was used to detect cell apoptosis of keloid fibroblasts. (d) Transwell assay was applied to assess the cell migration and invasion of keloid fibroblasts. (e) Western blot was applied to assess the levels of cyclinD1 and c-myc. (f) The mechanism diagram indicated that silencing circPTPN12 targets the miR-21-5p/SMAD7 axis and activates the Wnt signal pathway, thus affecting the progression of keloid. ** P < 0.01, *** P < 0.001.
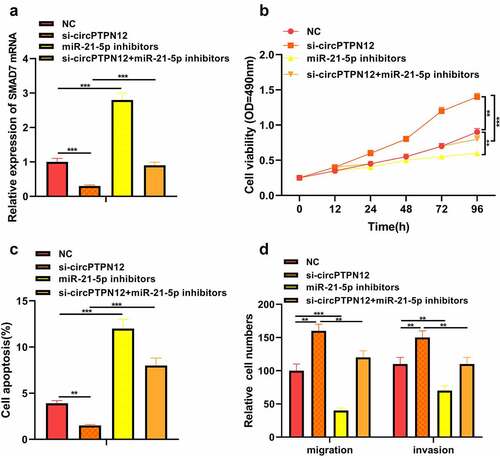
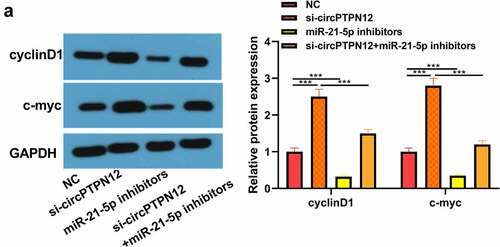
Figure 5. Silencing circPTPN12 targeted miR-21-5p/SMAD7 axis and activated the Wnt pathway to strengthen keloid fibroblasts’ growth. (a) The protein levels of cyclinD1 and c-myc were induced after silencing circPTPN12, while partly reversed by inhibition of miR-21-5p in keloid fibroblasts. Data were shown as mean ± SD.*** P < 0.001.
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
