Figures & data
Table 1. Primers and sequences used in the present research
Figure 1. Knockdown of LINC00665 suppressed cell proliferation in breast cancer cells.
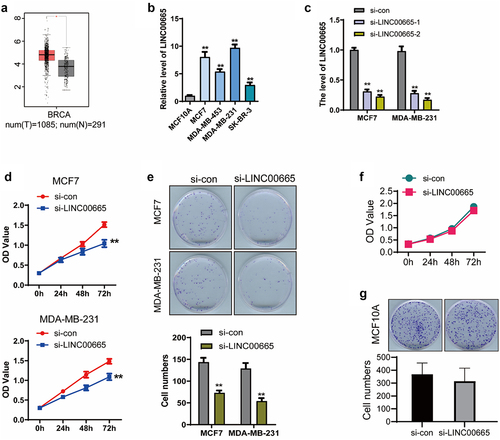
Figure 2. Knockdown of LINC00665 inhibited the migration and invasion and induced the apoptosis in breast cancer cells.
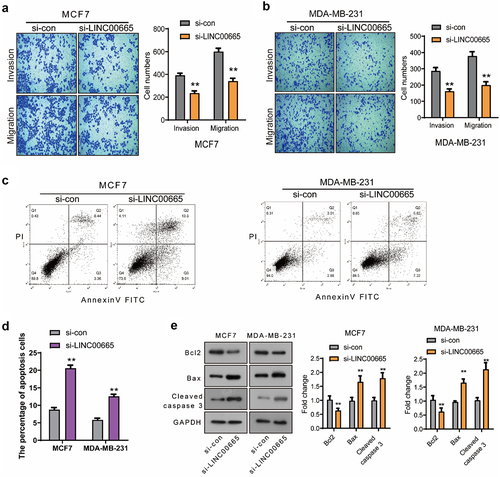
Figure 3. Knockdown of LINC00665 inactivated the AKT/mTOR signaling pathway in breast cancer cells.
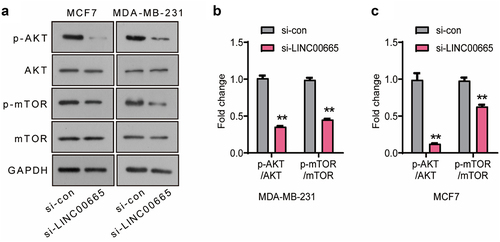
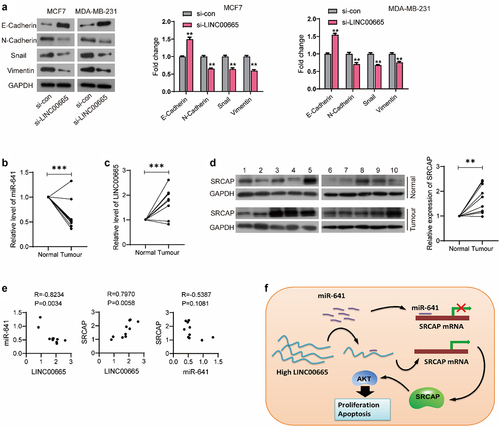
Figure 4. LINC00665 promoted the expression of SRCAP through sponging miR-641 in breast cancer cells.
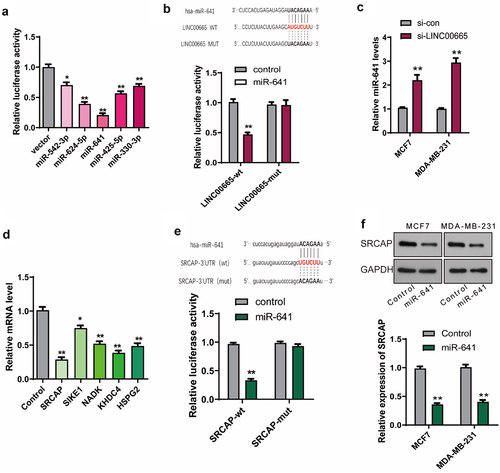
Figure 5. Knockdown of LINC00665 inhibited the proliferation and invasion through down-regulating SRCAP by sponging miR-641.
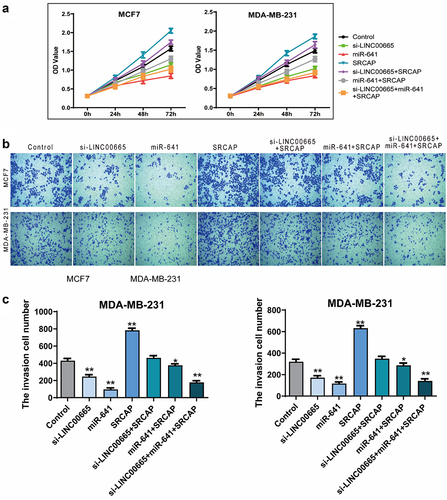
Figure 6. LINC00665 was up-regulated in breast cancer patients.