Figures & data
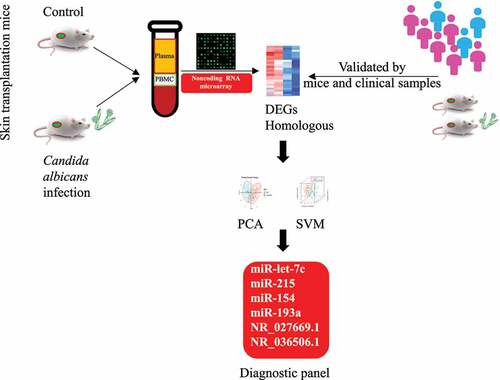
Figure 2. Discover differentially expressed miRNA and lncRNA in fungus-infected mice after transplant. (a) Heatmap of differentially expressed miRNAs in the plasma. (b) Heatmap of differentially expressed miRNAs in the peripheral blood mononuclear cell. (c) Heatmap of differentially expressed lncRNAs in the peripheral blood mononuclear cell.
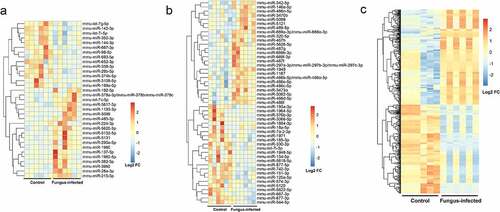
Figure 3. Identification of novel biomarkers for fungal infection after organ transplantation. (a) Real-time quantitative PCR validation of differentially expressed miRNAs and lncRNAs in the peripheral blood sample from a murine model of fungal infection. (b) Verification of miR-215 in plasma from transplanted patients with various fungal infections. (c) Validation of miR-let-7 c in plasma from transplanted patients with different kinds of fungus infection. (d) Expression of miR-154 in PBMC from transplanted patients with various fungal infections. (e) Validation of miR-193a in PBMC from transplanted patients with different kinds of fungus infection. (f) Verification of NR_027669.1 in PBMC from different fungus-infected patients after transplantation. (g) Expression of NR_036506.1 in PBMC from posttransplant patients with various fungal infections.
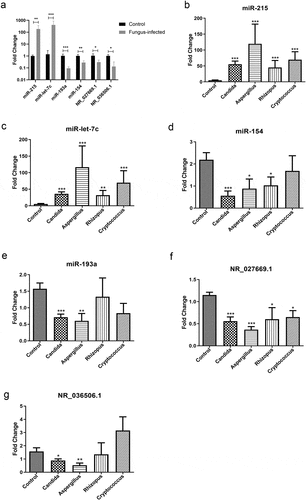
Table 1. The expression of different biomarkers in patients with various fungal infection sites
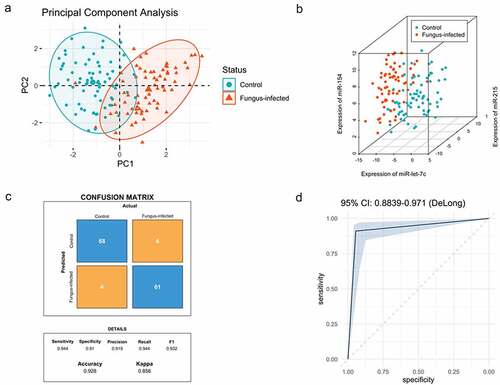
Figure 4. Expression pattern analysis of biomarkers from normal and fungal infected transplant recipients and classification model for normal and fungal infection (a) PCA analysis between normal and fungal infected patients after organ transplantation. (b) SVM analysis between normal and fungal infected patients after organ transplantation. (c) The confusion matrix method was used to test the model performance, and its accuracy, precision, sensitivity, and specificity were shown below. (d) The receiver operating characteristic (ROC) analysis was conducted with 2000 stratified bootstrap replicates.