Figures & data
Table 1. The difference in clinical data of the study population
Figure 1. DANCR levels in the serum of patients with atherosclerosis. (a) The serum DANCR was highly expressed in the serum of patients with atherosclerosis compared to healthy controls. (P < 0.001). (b) ROC curve analysis showed that DANCR could distinguish patients with atherosclerosis from healthy individuals. The AUC was 0.947, a sensitivity of 78.1%, and a specificity of 71%.
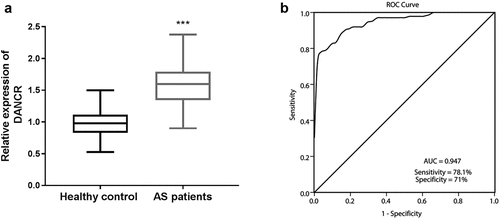
Table 2. The correlation analysis between DANCR expression and risk factors of atherosclerosis
Figure 2. Association between DANCR expression and LDL-C, Hcy, and CRP levels. (a) LDL-C levels were higher in the high DANCR expression group than the low DANCR expression group. (b) Hcy levels were high in the high DANCR expression group. (c) DANCR expression was positively associated with CRP levels. **P < 0.01, ***P < 0.001.
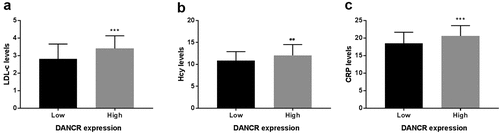
Figure 3. Regulation of DANCR on proliferation and migration of VSMC cells. (a) Expression of DANCR in VSMC cells transfected with siRNA or the negative control. (b) The proliferation of VSMC cells was measured using a CCK-8 assay after DANCR knockdown. (c) The migration of VSMC cells was detected through a Transwell migration assay after DANCR knockdown. (Scale bar, 50 μm) *P < 0.05, **P < 0.01, ***P < 0.001.
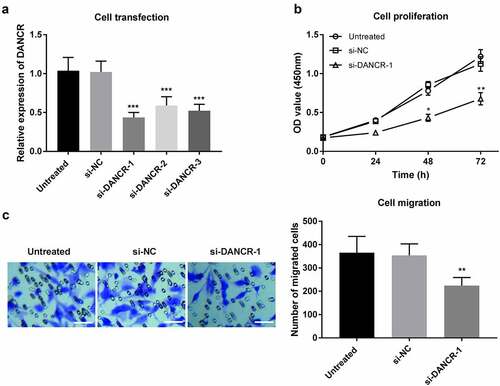
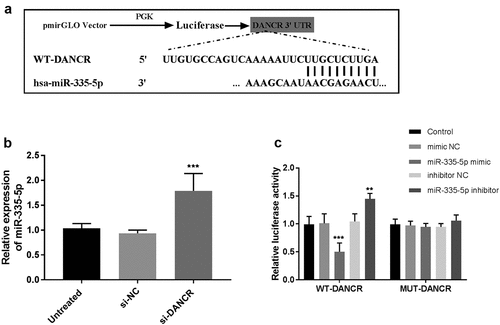
Figure 4. miR-335-5p is a target of DANCR in VSMC cells. (a) Bioinformatics analyses predicted the binding sites between DANCR and miR-335-5p. (b) The RT-qPCR was conducted to detect miR-335-5p expression in VSMC cells transfected with DANCR siRNA. (c) Dual-luciferase reporter assay validated the interaction between DANCR and miR-335-5p. **P < 0.01, ***P < 0.001.