Figures & data
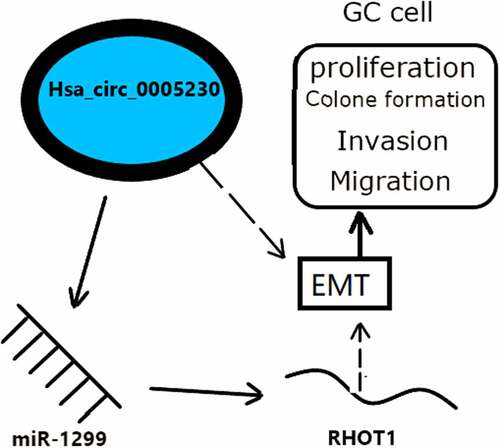
Table 1. Specific primers for qRT-PCR
Table 2. Relationship between different hsa_circ_0005230 expression and clinicopathological features of GC
Figure 1. The biological structure of hsa_circ_0005230 and its expression in GC tissues and cells.
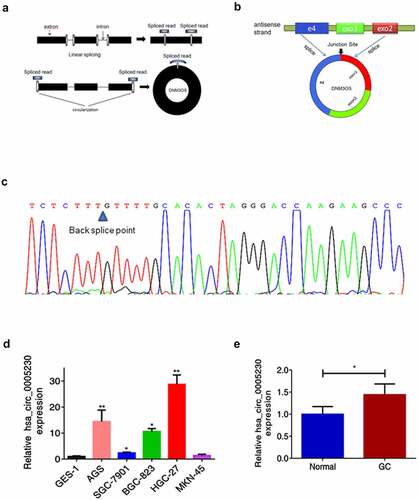
Figure 2. Silencing hsa_circ_0005230 not only diminished the capacities of clone formation and proliferation of GC cells but also arrested the cell cycle.
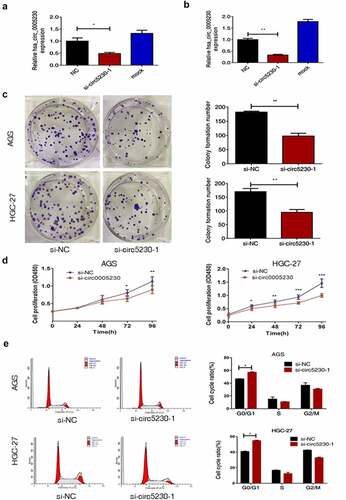
Figure 3. Silencing hsa_circ_0005230 inhibited the invasion and migration of GC cells.
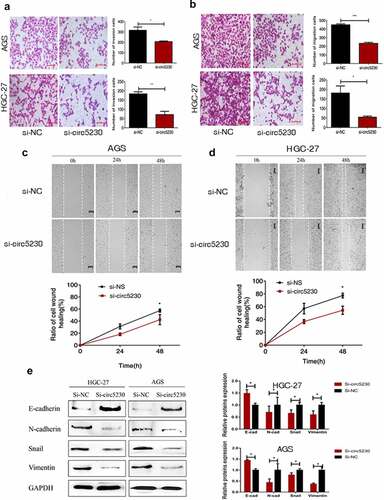
Figure 4. Hsa_circ_0005230 as a sponge to bind miR-1299.
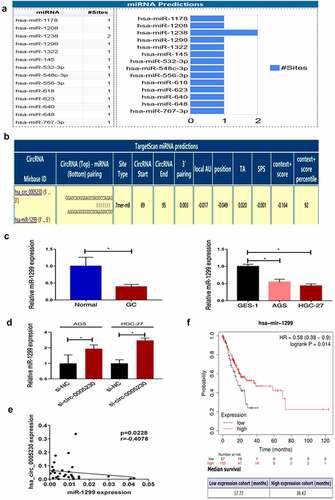
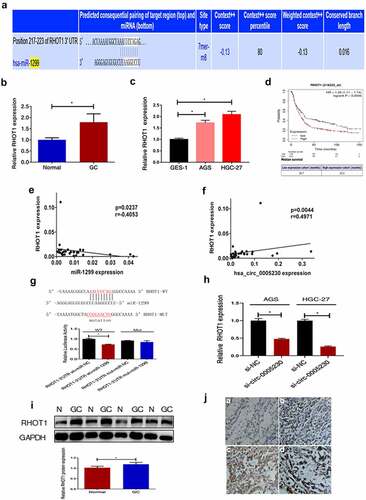
Figure 5. RHOT1 was the downstream target gene of the hsa_circ_0005230/miR-1299 axis.