Figures & data
Figure 1. Expressions of NEAT1 and miR-410-3p in normal and UC. (a) Expressions of NEAT1 and (b) miR-410-3p were examined by qRT-PCR in normal colon tissues and UC tissues. (c) IECs were transfected with control siRNA or NEAT1 siRNA for forty-eight hours, followed by treatments with LPS at the indicated concentrations for 24 hours. The cell responses were determined by MTT cell viability assay and (d) Annexin V apoptosis assay. *, p < 0.05.
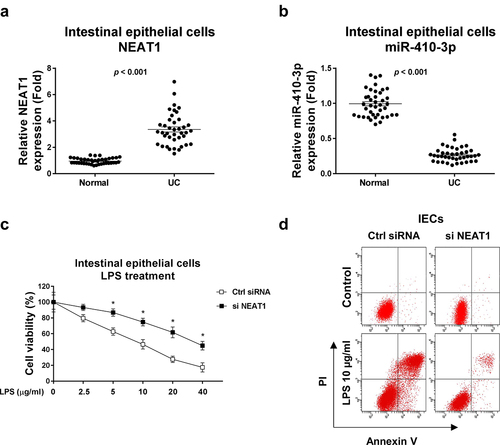
Figure 2. NEAT1 sponges miR-410-3p in IECs. (a) Prediction of the NEAT1-miR-410-3p association from starBase. (b) Correlation between NEAT1 and miR-410-3p expression in IECs from 40 UC tissues. (c) IECs were transfected with control or NEAT1 overexpression plasmid, expressions of miR-410-3p were determined by qRT-PCR. (d) RNA pull-down assay was performed in IECs by incubating cell lysates with the biotin-labeled control, sense or antisense NEAT1 probes. Amount of miR-410-3p in the RNA complex was assessed by qRT-PCR. (e) IECs isolated from normal and (f) UC tissues were transfected with WT-NEAT1 or Mut-NEAT1 plus control miRNA or miR-410-3p, luciferase reporter assay was conducted to examine the specific binding of miR-410-3p on NEAT1. **, p < 0.01; ***, p < 0.001.
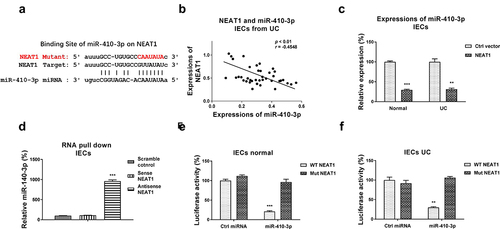
Figure 3. Glucose metabolism is elevated in IECs from UC. (a) Glucose uptake and (b) ECAR were detected in IECs from normal and UC tissues. (c) The glucose metabolism key enzymes, Hexokinase 2, LDHA and GLUT1 expressions were detected in IECs from normal and UC tissues by qRT-PCR. (d) IECs from normal and UC tissues were cultured under regular and low glucose conditions, the cell viability was assessed by clonogenic assay. **, p < 0.01; ***, p < 0.001.
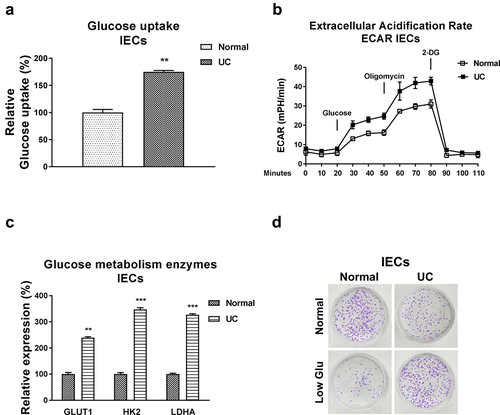
Figure 4. Reverse roles of NEAT1 and miR-410-3p in regulating glucose metabolism of IECs. (a) NEAT1 was overexpressed in IECs. (b) Glucose uptake (c) ECAR and (d) glucose metabolism key enzymes expressions from the above transfected cells were examined. (e) Control miRNA or miR-410-3p was transfected into IECs, expressions of miR-410-3p were examined by qRT-PCR. (f) Glucose uptake (g) ECAR and (h) glucose metabolism key enzymes expressions from the above transfected cells were examined. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
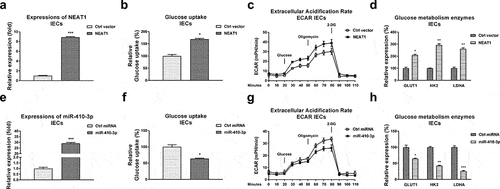
Figure 5. miR-410-3p targets LDHA to inhibit glucose metabolism. (a) Prediction of the binding sites of miR-410-3p on 3ʹUTR of LDHA from starBase. (b) Expressions of LDHA were examined in normal and UC tissues. (c) Correlation between miR-410-3p and LDHA in IECs from 40 UC tissues. (d) Control miRNA or miR-410-3p was transfected into IECs from normal and UC tissues. Protein expressions of LDHA were examined by Western blot. (e) IECs from normal and (f) UC tissues were transfected with 3UTR of WT-LDHA or Mut-LDHA plus control miRNA or miR-410-3p, luciferase reporter assay was conducted to examine the specific binding of miR-410-3p on LDHA. (g) IECs were transfected with control miRNA, miR-410-3p or miR-410-3p plus LDHA, expressions of LDHA were determined by Western blot. (h) Glucose uptake and (i) ECAR from the above transfected cells were measured. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
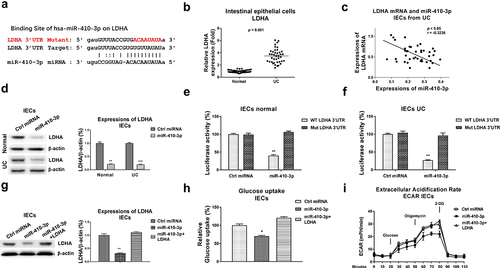
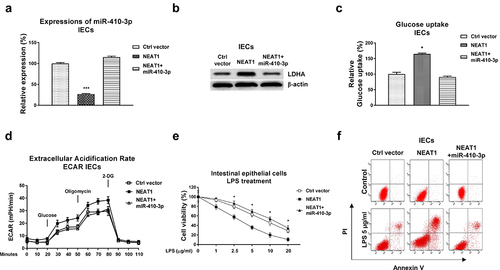
Figure 6. Roles of the NEAT1-miR-410-3p-LDHA in dysfunction of IECs. (a) IECs were transfected with control plasmid, NEAT1 alone or plus miR-410-3p, expressions of miR-410-3p and LDHA were determined by qPCR and (b) Western blot. (c) Glucose uptake and (d) ECAR from the above transfected cells were measured. (e) The above transfected cells were treated with LPS at the indicated concentrations. Cell viability and (f) Cell apoptosis were determined by MTT assay and Annexin V apoptosis assay. *, p < 0.05; ***, p < 0.001.