Figures & data
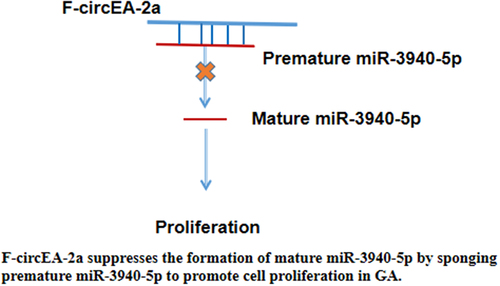
Figure 1. Accumulation of F-circEA-2a and miR-3940-5p (mature and premature) in GA Accumulation of F-circEA-2a, premature miR-3940-5p, and mature miR-3940-5p in both GA and non-tumor samples from GA patients (n = 70) was analyzed using RT-qPCR. Average values of three technical qPCR replicates were presented and compared. **, p < 0.01.
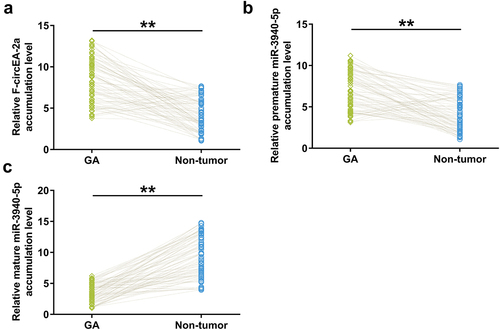
Figure 2. Correlation analysis between F-circEA-2a and miR-3940-5p (mature and premature) The correlation of F-circEA-2a with premature miR-3940-5p (a) or mature miR-3940-5p (b) was analyzed by Pearson’s correlation coefficient. Data presented in was used in this analysis.
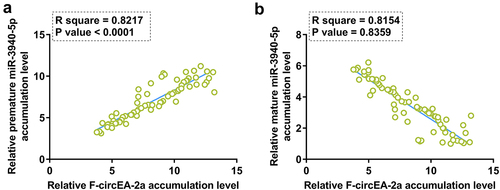
Figure 3. The binding of premature miR-3940-5p to F-circEA-2a The binding of premature miR-3940-5p to F-circEA-2a was predicted by IntaRNA 2.0 (a). The binding of premature miR-3940-5p to F-circEA-2a-wt (b) and F-circEA-2a-mut (B), which was labeled with red color in (A), was confirmed by RNA-RNA pull-down assay using biotin (Bio)-labeled RNAs. The enrichment of premature miR-3940-5p suggested the direct interaction between the two molecules. Data presented are the mean ± SD values of three biological replicates. **, p < 0.01.
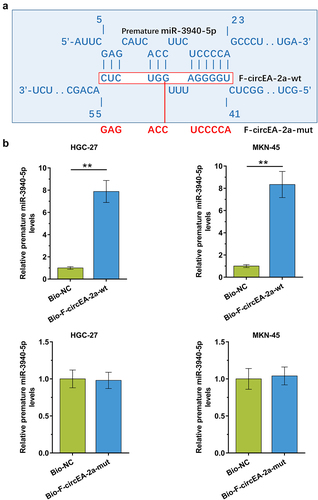
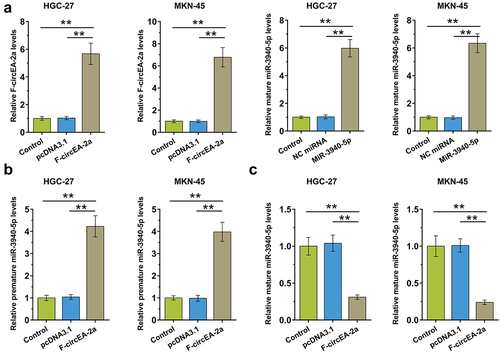
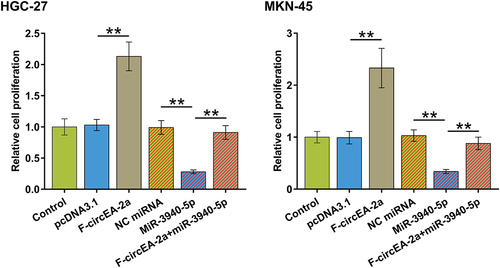
Figure 4. Regulation of miR-3940-5p maturation by F-circEA-2a HGC-27 and MKN-45 cells were overexpressed with F-circEA-2a and miR-3940-5p to explore their interaction (a). The role of F-circEA-2a in premature miR-3940-5p (b) and mature miR-3940-5p (c) accumulation was explored by RT-qPCR. Data presented are the mean ± SD values of three biological replicates. **, p < 0.01.