Figures & data
Table 1. The clinicopathological parameters in patients with CRC
Figure 1. LINC01315 is upregulated in CRC. a. Volcano maps of differentially expressed lincRNAs in GSE39582 and GSE41328. Red spots represent upregulated lincRNAs; green spots represent downregulated lincRNAs. b. Overlap of the dysregulated lincRNAs among GSE39582 and GSE41328. c. The expression of LINC01315 was analyzed using TCGA CRC dataset. d. LINC01315 expression level was significantly higher in CRC tissues than paracancerous tissues. **P < 0.01 compared with paracancerous. e. The effect of LINC01315 expression on clinical prognosis was determined by Kaplan-Meier survival analysis. The median value (0.65) was the cutoff threshold of Kaplan-Meier survival analysis. f. Expression level of LINC01315 was significantly higher in CRC cell lines than the human colon immortalized cell line, FHC. **P < 0.01 compared with FHC. g. The subcellular localization of LINC01315 predicted on lncATLAS website (https://lncatlas.crg.eu/).
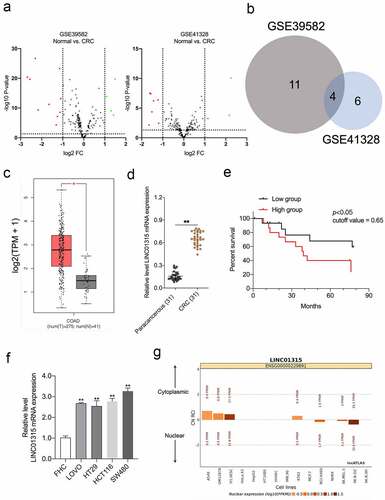
Figure 2. Deletion of LINC01315 inhibits CRC cells growth, invasion, and EMT. a. LINC01315 knockdown by shRNA (shLINC01315) was confirmed using qRT-PCR. b. CCK-8 assay showing proliferation of HCT116 and SW480 cancer cells following LINC01315 knockdown. c. Colony formation assays were performed to determine the proliferation of shLINC01315-transfected or shNC- transfected CRC cells. d. Wound-healing assay of LINC01315 knockdown HCT116 and SW480 cells. e. Transwell invasion assay analyzed the effect of LINC01315 knockdown on CRC cells invasion. f. Western blot assay was performed to test the expression of EMT markers (E-cadherin and N-cadherin) in the LINC01315 knockdown cells. **P < 0.05 compared with shNC.
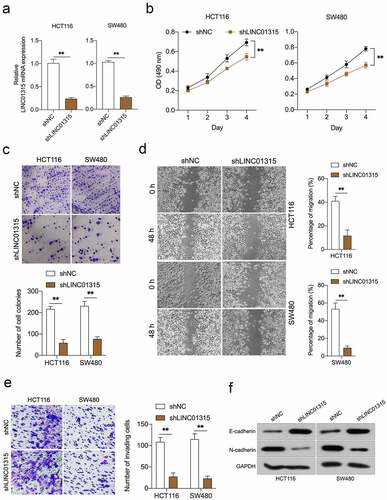
Figure 3. LINC01315 activates the Wnt/β-catenin pathway. a. LINC01315 knockdown results in a reduction of β-catenin expression. b. TOP/FOP luciferase reporter assays were performed to detect the β-catenin activity. c. Altered nuclear translocation of β-catenin in response to LINC01315 overexpression. d. Upregulation of LINC01315 raised the RNA levels on Wnt/β-catenin direct-target genes. **P < 0.05 compared with vector.
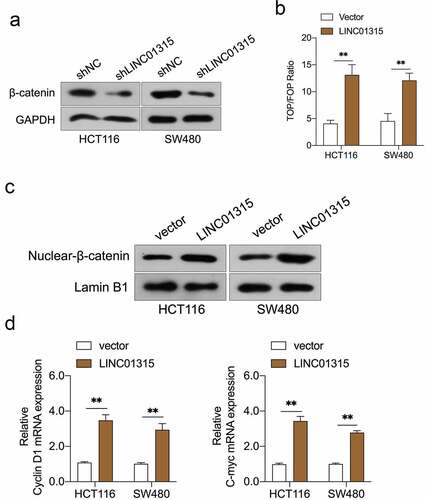
Figure 4. LINC01315 binds to the β-catenin promoter. a. pcDNA3.1-LINC01315 transfection increased β-catenin mRNA level. b. Relative luciferase activities of the β-catenin reporter in LINC01315 overexpressing CRC cells. c. The targeting relation of LINC01315 and β-catenin was confirmed by RNA pull-down assay. d. RIP was performed to measure the binding abilities of LINC01315 and β-catenin. e. The activity of the β-catenin promoter was measured by luciferase assay. **P < 0.05 compared with vector.
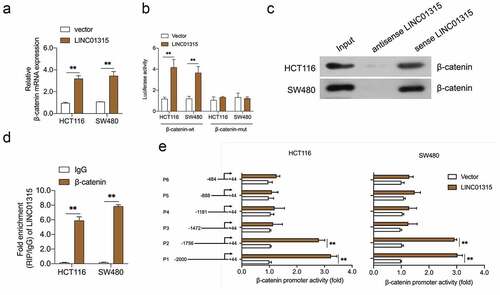
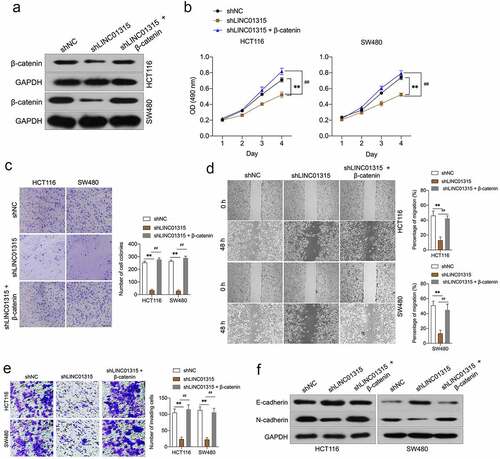
Figure 5. Overexpression of β-catenin reverses the effects of LINC01315 on CRC cells EMT. a. HCT116 and SW480 cells were co-transfected with shLINC01315 and pcDNA3.1-β-catenin. The expression of β-catenin was determined by Western blot. b. CCK-8 assay showing proliferation of HCT116 and SW480 cells. C. Colony formation assays were performed to determine the proliferation of shLINC01315 and pcDNA3.1-β-catenin co-transfected CRC cells. d. Wound-healing assay of HCT116 and SW480 cells. e. Transwell invasion assay analyzed CRC cells invasion. f. Western blot assay was performed to test the expression of EMT markers (E-cadherin and N-cadherin) in HCT116 and SW480 cells. **P < 0.05 compared with shNC, ##P < 0.05 compared with shLINC01315.
Data availability statement
The datasets used or/and analyzed during the current study are available from the corresponding author on reasonable request.
