Figures & data
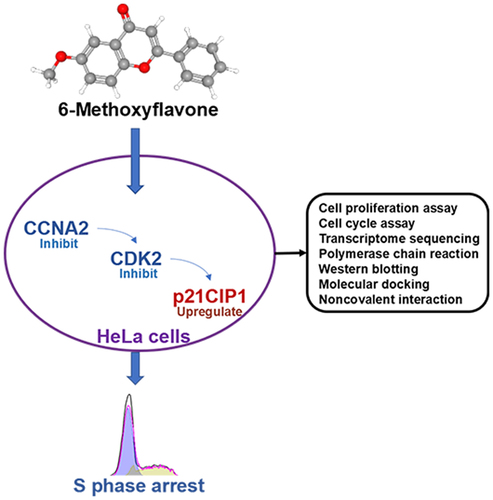
Table 1. Primer sequences used for the quantitative polymerase chain reaction
Figure 1. Functional enrichment analyses of 178 targets. A. The 10 most common KEGG pathways (p < 0.0001). B. KEGG pathways related to cancer. C. Biological processes related to cell proliferation. D and F. Functional enrichment terms related to cell cycles and drugs (p < 0.05). E. Genes enriched in six cell cycle items. G. Principal component analysis of the genes enriched in the cell cycle. H. Intersections of the 178 targets and the DEmRNAs of cervical cancer. I. Intersections among the targets of the principal component analysis and DEmRNAs. Abbreviations: KEGG: Kyoto Encyclopedia of Genes and Genomes; DEmRNAs: differentially expressed mRNAs; FoxO: forkhead box O; PPAR: peroxisome proliferator activated receptor; CCNA2: cyclin A2; DHFR: dihydrofolate reductase; CHEK1: checkpoint kinase 1; EGFR: epidermal growth factor receptor; ADAM17: ADAM metallopeptidase domain 17; CDK2: cyclin-dependent kinase 2; CDK6: cyclin dependent kinase 6; CDK7: cyclin-dependent kinase 7; GSK3B: glycogen synthase kinase 3 beta; TGFB2: transforming growth factor beta 2; EIF4E: eukaryotic translation initiation factor 4E; PIM1: Pim-1 proto-oncogene, serine/threonine kinase; FGFR1: fibroblast growth factor receptor 1; FGFR2: fibroblast growth factor receptor 2.
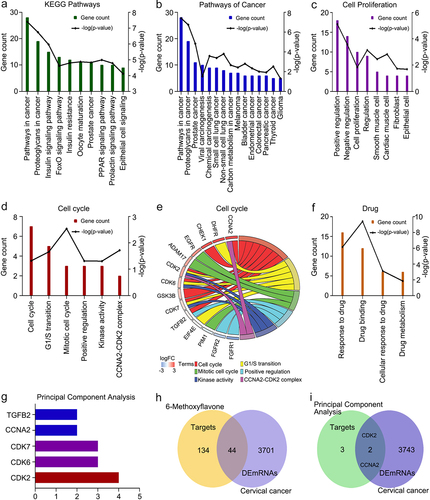
Figure 2. Gene set enrichment analyses of CCNA2 and CDK2. A. The five most common KEGG pathways or biological processes of each dataset (q < 0.01). B. The highest normalized enrichment score terms of each dataset. Abbreviations: KEGG: Kyoto Encyclopedia of Genes and Genomes; P53: tumor protein p53; CCNA2: cyclin A2; CDK2: cyclin-dependent kinase 2.
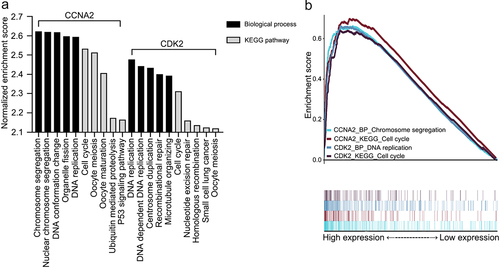
Figure 3. Eukaryotic transcriptome sequencing results of HeLa cells after 6-methoxyflavonoid treatment. Three control samples were treated with 0.16% dimethyl sulfoxide. Three treatment samples were treated with 65 μM 6-methoxyflavone. A. Minus versus add plot of differential expressed transcripts. B. The 10 most common KEGG pathways of differential expressed transcripts (p < 0.05). C and D. Gene oncology enrichment results of differential expressed transcripts. Cell cycle-related or proliferation-related biological processes (p < 0.05). Abbreviations: TPM: transcripts per million; HTLV-I: human T-lymphotropic virus type I; PI3K: phosphoinositide 3-kinase; Akt: protein kinase B.
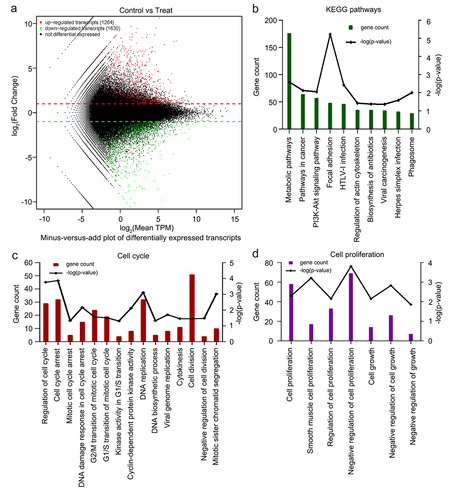
Figure 4. 6-Methoxyflavone inhibits cervical cancer cell proliferation and induces S-phase arrest in HeLa cells. A. Inhibition ratios of HaCaT, HeLa, C33A, and SiHa cells. The four cells were treated with six concentrations of 6-methoxyflavone (0.16% DMSO, 20 μM, 40 μM, 80 μM, 120 μM, and 160 μM) for 48 hours. Six replicate holes were used at each concentration level. A water-soluble tetrazolium salt-8 kit was used to measure the inhibitory effects. B. Inhibition ratio of HeLa cell proliferation. HeLa cells were treated with six concentrations of 6-methoxyflavone for 24 hours, 48 hours, and 72 hours. C. After propidium iodide staining, the distribution of HeLa cell cycle phases was measured using flow cytometry. D. The histogram shows the ratio of the HeLa cell cycle phases in . All experiments were performed using three biological replicates. Statistical analysis was performed using a one-way analysis of variance (ANOVA). *p < 0.05. **p < 0.01. n.s.: not significant. DMSO: dimethyl sulfoxide.
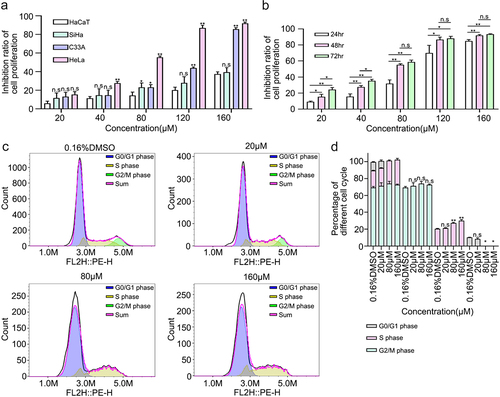
Table 2. Core mRNA biomarker expression levels of transcriptome sequencing
Figure 5. 6-Methoxyflavone induces S-phase arrest through the CCNA2/CDK2/p21CIP1 pathway. HeLa cells were treated with 6-methoxyflavone (0.16% DMSO and 65 μM) for 48 hours. Cell cycle-related mRNA and protein expression were measured by PCR and western blotting. A. The relative mRNA expressions of CCNA2, CCNB1, CCND1, CCNE1, CDK1, CDK2, CDK4, CDK6, P16INK4, p21CIP1, TP53, and RB. B and C. Protein expressions of CCNA2, CCND1, CCNE1, CDK2, CDK6, and p21CIP1. Each assay was performed in triplicate. Statistical analysis was performed using the paired t-test. *p < 0.05. **p < 0.01. n.s.: not significant. Abbreviations: CCNA2: cyclin A2; CCNB1: cyclin B1; CCND1: cyclin D1; CCNE1: cyclin E1; CDK1: cyclin-dependent kinase 1; CDK2: cyclin-dependent kinase 2; CDK4: cyclin-dependent kinase 4; CDK6: cyclin dependent kinase 6; P16INK4: cyclin dependent kinase inhibitor 2A; p21CIP1: cyclin-dependent kinase inhibitor 1A; TP53: tumor protein p53; RB: retinoblastoma transcriptional corepressor 1.
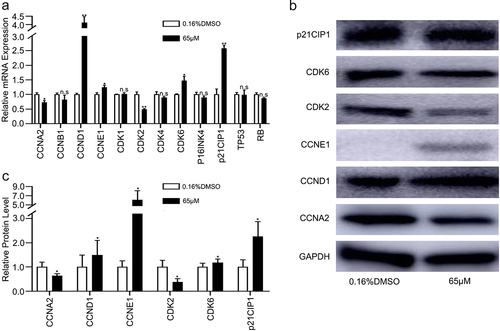
Table 3. Molecular docking parameters of 6-methoxyflavone and receptors
Figure 6. Affinity and noncovalent interactions between 6-methoxyflavone and receptors. A and C. The results of molecular dockings between 6-methoxyflavone and CCNA2, CDK2, p21CIP1, and the CCNA2–CDK2 complex. B and D. The noncovalent interactions between 6-methoxyflavone and CCNA2, CDK2, and CCNA2–CDK2 complex. E. The noncovalent interactions between 6-methoxyflavone and CCNA2, CDK2, and the CCNA2–CDK2 complex. Abbreviations: CCNA2: cyclin A2; CDK2: cyclin-dependent kinase 2; p21CIP1: cyclin-dependent kinase inhibitor 1A.
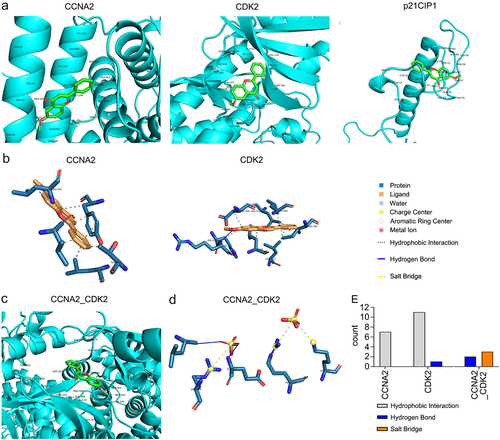
Table 4. Relationships between mRNA expression levels of six targets and the clinicopathological parameters of cervical cancer patients
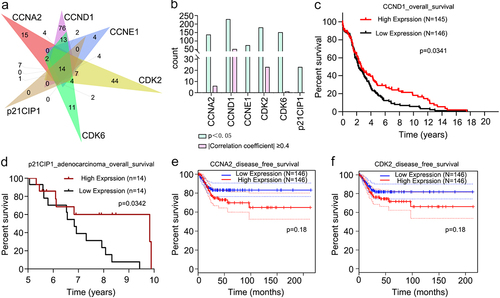
Figure 7. Clinical characteristics analyses of six receptors. A. Drug sensitivity data of CCNA2, CCND1, CCNE1, CDK2, CDK2, and p21CIP1. Numbers of drugs related to the six receptors (p < 0.05). B. Drugs related to the six receptors (p < 0.05). Drugs significantly related to the expressions of CCNA2, CCND1, CDK2, and CDK6 (p < 0.05 and |Pearson and Spearman correlation coefficient|≥0.4). C. Overall survival analysis of cervical cancer patients in CCND1 high-expression and low-expression groups (p < 0.05). D. Overall survival analysis of cervical adenocarcinoma patients in p21CIP1 high-expression and low-expression groups(p < 0.05). E and F. Disease free survival analyses of cervical cancer patients in CCNA2 and CDK2 different expression groups.