Figures & data
Table 1. Correlation analysis for circ_0002360 expression and the clinicopathological parameters of patients with NSCLC (n = 68)
Figure 1. The expression of circ_0002360 was increased in Taxol-resistant NSCLC. (a) GSE158695 and GSE112214 datasets were used to analyze the expression of circ_0002360 in NSCLC tissues. (b) The expression of circ_0002360 in NSCLC tissues and normal tissues was detected by qRT-PCR. (c) The expression of circ_0002360 in Taxol-sensitive and resistant tissues of NSCLC was detected by qRT-PCR. (d) The IC50 values of Taxol in H1299/Taxol and A549/Taxol cells were tested via CCK-8 assay. (e) The expression of circ_0002360 in normal cells, Taxol-sensitive and resistant cells of NSCLC was assessed. (f and g) The abundance of circ_0002360 and RUNX1 mRNA levels after treatment with RNase R. (h) The existence of circ_0002360 was detected in cell lines by qRT-PCR using cDNA or gDNA as templates with divergent or convergent primers and verified by Gel electrophoresis. ◄►: divergent primers, ►◄: convergent primers. (i) Schematic diagram showed the formation of circ_0002360. ***P < 0.001, ****P < 0.0001.
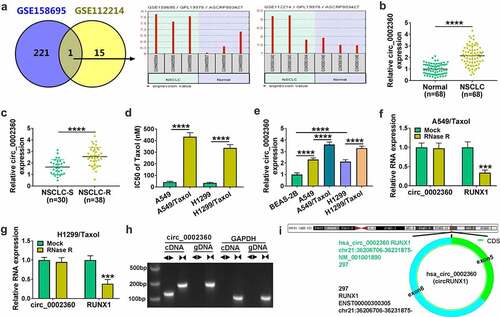
Figure 2. Circ_0002360 knockdown regulated the behaviors of Taxol-resistant cells. (a) The transfection efficiency of sh-circ_0002360 in H1299/Taxol and A549/Taxol cells. (b) IC50 value of Taxol in cells transfected as indicated by CCK-8. (c) Colony formation assay performed with cells transfected as indicated. (d) EdU assay for proliferation of cells transfected as indicated. (e) The activity of caspase-3 in cells transfected as indicated using an assay kit. (f) Flow cytometry performed with cells transfected as indicated to evaluate apoptosis. (G to I) The ability of cells transfected as indicated to migrate and invade was detected by the Transwell assay. (j) PCNA, Bax, MRP1 and P-gp levels in cells transfected as indicated. **P < 0.01, ***P < 0.001, ****P < 0.0001.
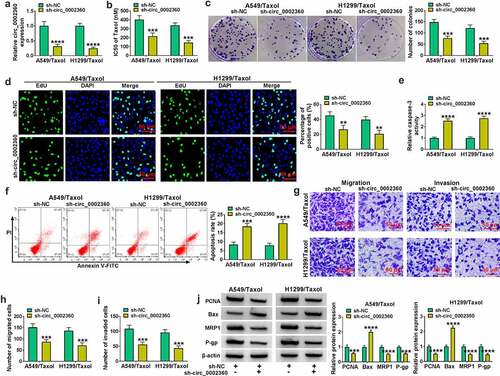
Figure 3. MiR-585-3p was targeted by circ_0002360. (a) The localization of circ_0002360 in cells was detected by subcellular localization experiment. (b) Predicted duplex formation between circ_0002360 and miR-585-3p by CircInteractome. (c and d) Dual-luciferase report assay in H1299/Taxol and A549/Taxol cells. (d) RIP assay in H1299/Taxol and A549/Taxol cells. (e) MiR-585-3p expression in LUAD samples and healthy samples in and LUSC. (f) MiR-585-3p level in NSCLC samples and normal samples. (g) MiR-585-3p level in Taxol-resistant and sensitive tissues in NSCLC. (h) Correlation analysis between circ_0002360 and miR-585-3p expression in NSCLC tissues. (i) MiR-585-3p level in normal cells, NSCLC cells and Taxol-resistant cells in NSCLC. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
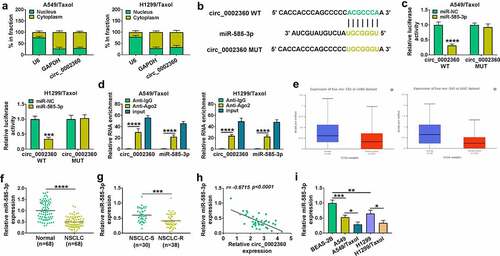
Figure 4. Anti-miR-585-3p reversed the impact of sh-circ_0002360 in H1299/Taxol and A549/Taxol cells. (a) MiR-585-3p level in H1299/Taxol and A549/Taxol cells after transfection by sh-NC, sh-circ_0002360, sh-circ_0002360+ anti-NC or sh-circ_0002360+ anti-miR-585-3p. (b) IC50 value of Taxol in cells transfected as indicated by CCK-8. (c) Colony formation assay performed with cells transfected as indicated. (d) EdU assay for proliferation of cells transfected as indicated. (e) The activity of caspase-3 in cells transfected as indicated using an assay kit. (f) Flow cytometry performed with cells transfected as indicated to evaluate apoptosis. (g and h) The ability of cells transfected as indicated to migrate and invade was detected by the Transwell assay. (i) PCNA, Bax, MRP1 and P-gp levels in cells transfected as indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
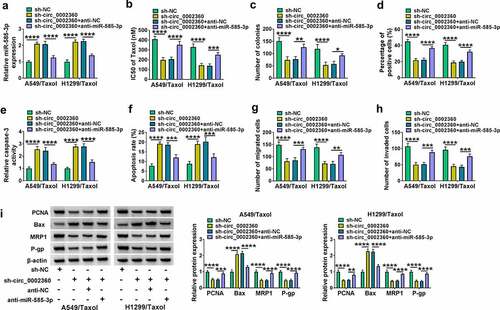
Figure 5. GPRIN1 was targeted by miR-585-3p. (a) Predicted duplex formation between miR-585-3p in GPRIN1. (b) Dual-luciferase reporting assay in H1299/Taxol and A549/Taxol cells. (c) GPRIN1 mRNA level in LUAD and LUSC. (d) GPRIN1 mRNA level in NSCLC samples compared with normal human samples. (e) GPRIN1 level in Taxol-sensitive and resistant samples. (f) Correlation analysis between GPRIN1 and miR-585-3p expression. (g) Correlation analysis between circ_0002360 expression and GPRIN1 expression. (h) GPRIN1 protein level in Taxol-resistant and sensitive samples. (i) GPRIN1 protein level in normal cells, NSCLC cells and Taxol-resistant cells in NSCLC. (J to L) GPRIN1 protein level in H1299/Taxol and A549/Taxol cells after miR-585-3p or anti-miR-585-3p transfection. (m and n) GPRIN1 protein level in cells transfected as indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
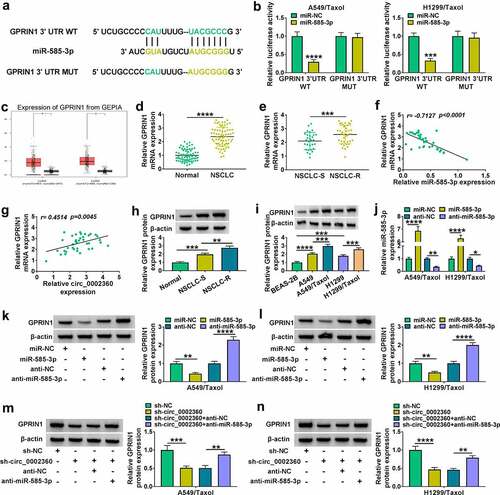
Figure 6. GPRIN1 overexpression reversed the influence of miR-585-3p in H1299/Taxol and A549/Taxol cells. (a) GPRIN1 level in H1299/Taxol and A549/Taxol cells after transfection by vector and GPRIN1. (b-i) H1299/Taxol and A549/Taxol cells were introduced with miR-NC mimics, miR-585-3p mimics, miR-585-3p mimics+vector or miR-585-3p mimics+GPRIN1. (b) IC50 value of Taxol in cells transfected as indicated by CCK-8. (c) Colony formation assay performed with cells transfected as indicated. (d) EdU assay for proliferation of cells transfected as indicated. (e) The activity of caspase-3 in cells transfected as indicated using an assay kit. (f) Flow cytometry performed with cells transfected as indicated to evaluate apoptosis. (g and h) The ability of cells transfected as indicated to migrate and invade was detected by the Transwell assay. (i) PCNA, Bax, MRP1 and P-gp levels in cells transfected as indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
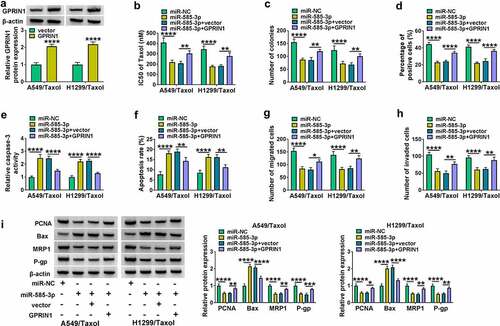
Figure 7. Circ_0002360 inhibited the growth of NSCLC cells and enhanced their sensitivity to Taxol in vivo. (a) Tumor volume was periodically monitored. (b and c) Images and average weight of the xenograft tumors in each group at the endpoint. (d) The expression of circ_0002360, miR-585-3p and GPRIN1 in each group. (e) Western blot for GPRIN1, MRP1 and P-gp levels in xenograft tumors. (f) PCNA staining cells in xenograft tumors detected by IHC. **P < 0.01, ***P < 0.001, ****P < 0.0001.
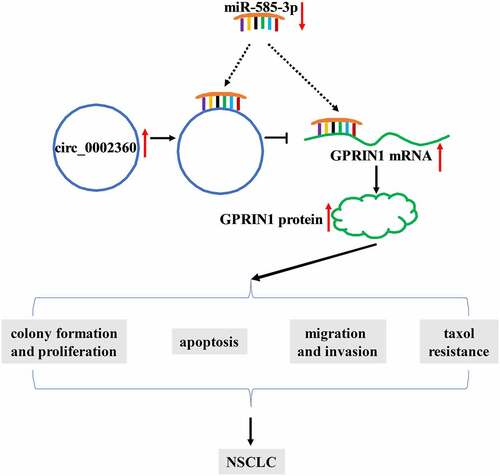
Figure 8. Schematic model of the circ_0002360/miR-585-3p/GPRIN1 axis in NSCLC Taxol resistance. In Taxol-resistant NSCLC cells, circ_0002360 was upregulated and miR-585-3p was underexpressed and thus GPRIN1 expression was enhanced, thereby promoting Taxol resistance, cell proliferation, metastasis and diminishing cell apoptosis.