Figures & data
Figure 1. LINC01270 expression was up-regulated in GC.
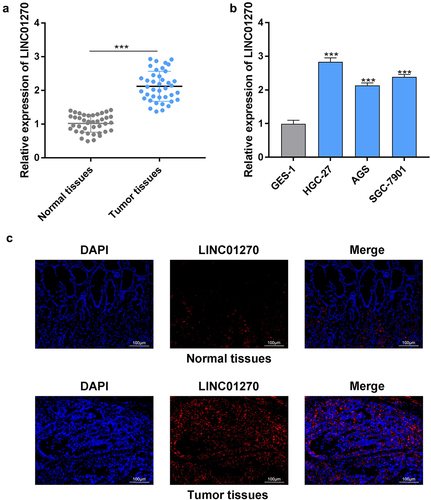
Figure 2. LINC01270 knockdown retarded GC progression.
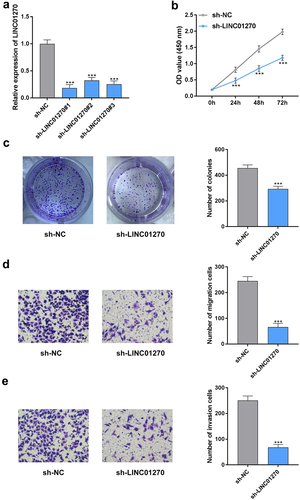
Figure 3. LINC01270 suppression reduced tumor growth in vivo.
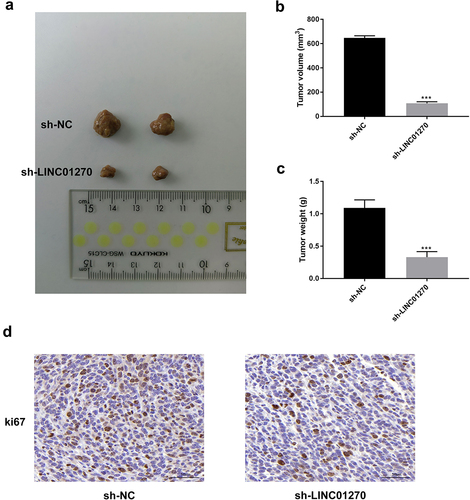
Figure 4. LINC01270 sponged miR-326.
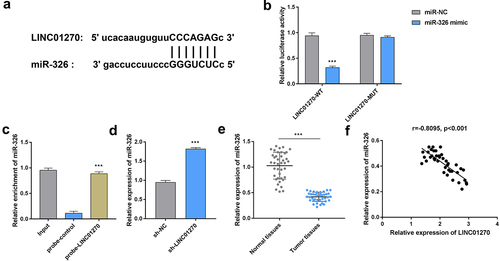
Figure 5. MiR-326 targeted and regulated EFNA3.
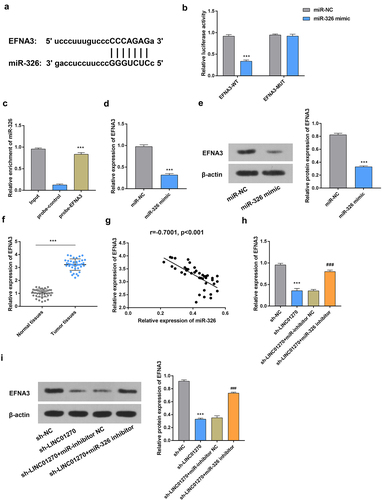
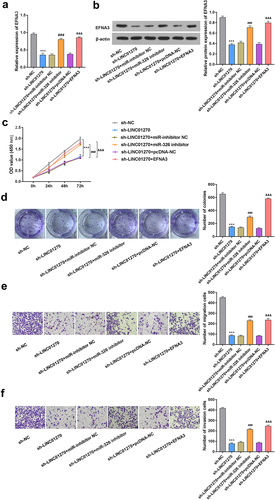
Figure 6. LINC01270 affected GC progression via miR-326/EFNA3 axis.
Supplemental Material
Download Zip (2.5 MB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
