Figures & data
Table 1. Primers sequences used for qRT-PCR
Figure 1. The expression of circ_0086414 in EC tissues and cells. (a) GEO dataset was used to analyze differently expressed circRNAs through bioinformatics method. (b) Circ_0086414 expression was analyzed in EC tissues and normal esophageal tissues through the GEO dataset (GSE131969). (c) Circ_0086414 expression was checked by qRT-PCR in 53 pairs of EC tissues and paracancerous normal esophageal tissues. (d) Kaplan-Meier method was carried out to analyze the association between circ_0086414 expression and overall survival of EC patients. (e) Circ_0086414 expression was checked by qRT-PCR in Het-1A, KYSE150, TE-10, KYSE450 and TE-1 cells. (f and g) Subcellular fractionation location assay was used to demonstrate that circ_0086414 was mainly located in the cytoplasm. (h and i) The stability of circ_0086414 was identified using random primers and Oligo(dT)18 primers. *p< 0.05.
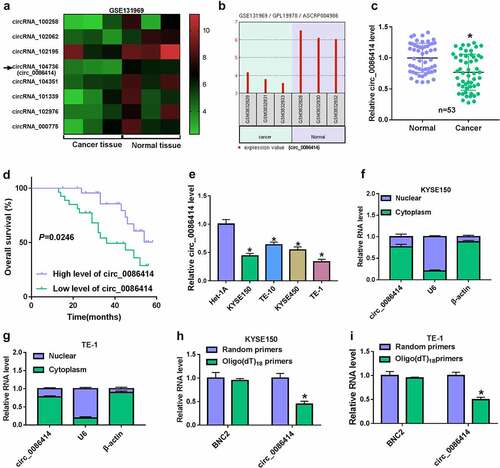
Figure 2. The effects of circ_0086414 on EC cell malignancy. (a) The efficiency of circ_0086414 overexpression was determined by qRT-PCR. (b-k) KYSE150 and TE-1 cells were transfected with pCD5-ciR or circ_0086414, respectively, and cell proliferation was investigated by MTT, EdU and cell colony formation assays (b-e), cell invasion by transwell invasion assay (f), the protein expression of N-cadherin and E-cadherin by Western blotting analysis (g and h), glucose consumption by glucose assay kit (i), lactate production by lactate assay kit (j), and cell apoptosis by flow cytometry analysis (k). *P< 0.05.
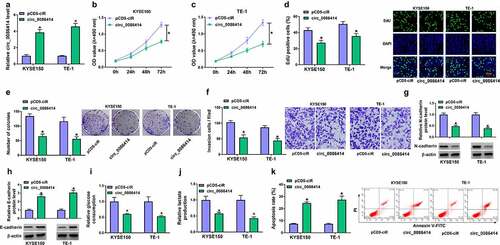
Figure 3. Ectopic expression of circ_0086414 delayed tumor tumorigenesis. (a-d) The effects of circ_0086414 overexpression on tumor volume and weight. (e) Circ_0086414 expression was detected by qRT-PCR in the forming tumors from the pCD5-ciR or circ_0086414 group. *P< 0.05.
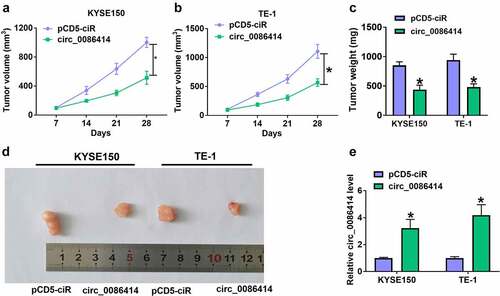
Figure 4. Circ_0086414 acted as a sponge for miR-1290. (a) The diagram illustration showing the complementary sites of circ_0086414 with miR-1290. (b-d) Dual-luciferase reporter and RNA pull-down assays were performed to identify the interaction of circ_0086414 and miR-1290 in KYSE150 and TE-1 cells. (e and f) MiR-1290 expression in EC tissues (N = 119) and normal esophageal tissues (N = 119) was analyzed through bioinformatics method. (g and i) MiR-1290 expression was detected by qRT-PCR in 53 pairs of EC and paracancerous normal esophageal tissues, and het-1A, TE-10, KYSE150, KYSE450 and TE-1 cells. (h) The correlation of circ_0086414 expression and miR-1290 expression in EC tissues was analyzed by spearman correlation analysis. (j) The effect of circ_0086414 overexpression on miR-1290 expression level was determined by qRT-PCR in both KYSE150 and TE-1 cells. *P< 0.05.
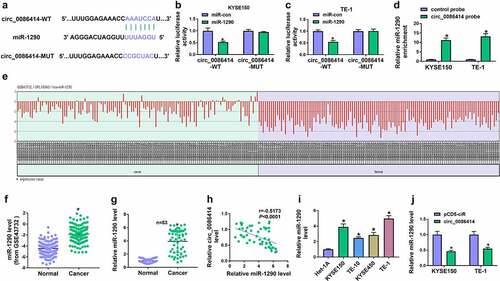
Figure 5. The effects of circ_0086414 and miR-1290 on EC cell malignancy. (a) The efficiency of miR-1290 overexpression was determined by qRT-PCR. (b-t) KYSE150 and TE-1 cells were transfected with pCD5-ciR, circ_0086414, circ_0086414+ miR-con, or circ_0086414+ miR-1290, respectively, and miR-1290 expression was analyzed by qRT-PCR (b), cell proliferation was investigated by MTT, EdU and cell colony formation assays (c-h), cell invasion by transwell invasion assay (i and j), the protein expression of N-cadherin and E-cadherin by Western blotting analysis (k-n), glucose consumption by glucose assay kit (o and p), lactate production by lactate assay kit (q and r), and cell apoptosis by flow cytometry analysis (s and t). *P< 0.05.
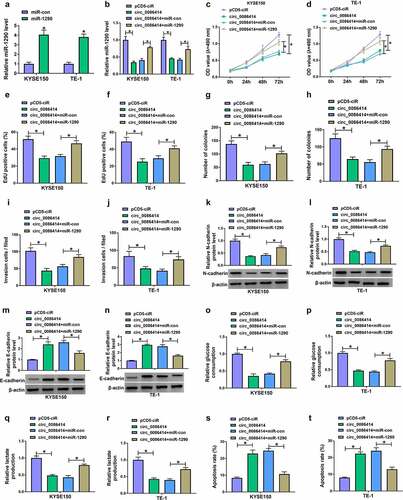
Figure 6. MiR-1290 targeted SPARCL1 in KYSE150 and TE-1 cells. (a) The diagram illustration showing the binding sites of miR-1290 for SPARCL1. (b-d) Dual-luciferase reporter and RNA pull-down assays were carried out to analyze the interaction between miR-1290 and SPARCL1 in KYSE150 and TE-1 cells. (e) SPARCL1 expression was analyzed using the TCGA dataset (GEPIA) in EC tissues and normal esophageal tissues. (f) SPARCL1 mRNA expression was detected by qRT-PCR in 53 pairs of EC tissues and normal esophageal tissues. (g) The correlation of miR-1290 expression and SPARCL1 expression in EC tissues was analyzed by spearman correlation analysis. (h and i) Western blotting analysis was performed to detect SPARCL1 protein expression in EC tissues, normal esophageal tissues, and Het-1A, TE-10, KYSE150, KYSE450 and TE-1 cells. (j) The efficiency of miR-1290 knockdown was determined by qRT-PCR in KYSE150 and TE-1 cells. (k) The effect of miR-1290 depletion on SPARCL1 protein expression was analyzed by western blotting analysis in KYSE150 and TE-1 cells. *P< 0.05.
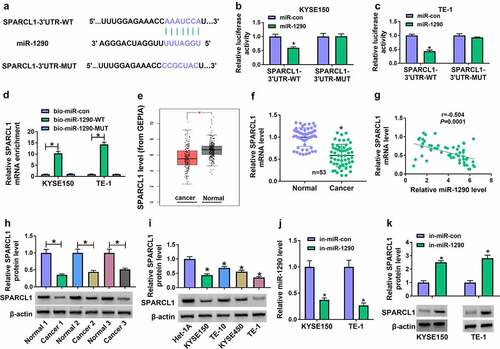
Figure 7. MiR-1290 regulated EC cell processes through SPARCL1. (a) The efficiency of SPARCL1 knockdown was determined by western blotting analysis in KYSE150 and TE-1 cells. (b-t) KYSE150 and TE-1 cells were transfected with in-miR-con, in-miR-1290, in-miR-1290+ si-con or in-miR-1290+ si-SPARCL1, and SPARCL1 protein expression was analyzed by western blotting analysis (b), cell proliferation by MTT, EdU and cell colony formation assays (c-h), cell invasion by transwell invasion assay (i and j), the protein expression of N-cadherin and E-cadherin by western blotting analysis (k-n), glucose consumption by glucose assay kit (o and p), lactate production by lactate assay kit (q and r), and cell apoptosis by flow cytometry analysis (s and t). *P< 0.05.
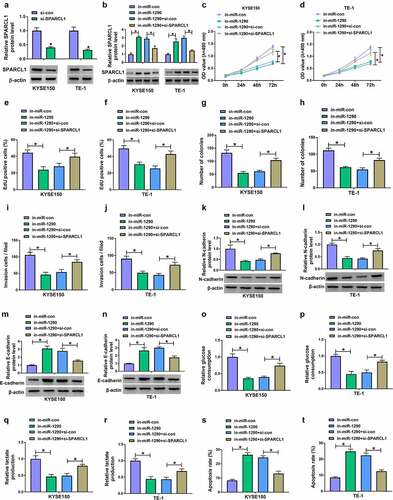
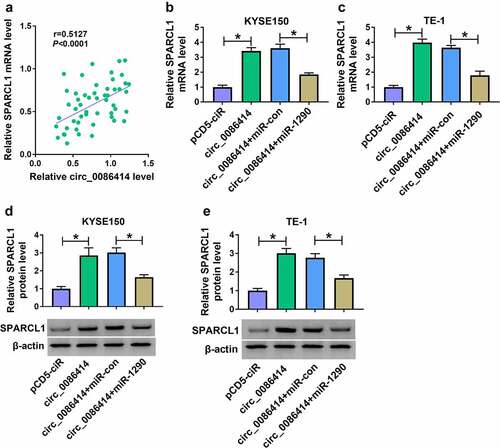
Figure 8. Circ_0086414 regulated SPARCL1 expression by interacting with miR-1290. (a) The correlation of circ_0086414 and SPARCL1 expression in EC tissues was analyzed by spearman correlation analysis. (b-e) KYSE150 and TE-1 cells were transfected with pCD5-ciR, circ_0086414, circ_0086414+ miR-con or circ_0086414+ miR-1290, respectively, and SPARCL1 mRNA expression was detected by qRT-PCR (b and c), and SPARCL1 protein expression by western blotting analysis (d and e). *P< 0.05.