Figures & data

Table 1. Primers used for the target genes
Figure 1. Dex pretreatment promotes the viability of H9C2 cells under H/R and inhibits apoptosis and inactivate Wnt/β-catenin pathway (a). The cell viability in H9C2 treated with different concentrations of Dex (0, 1, 5, 10 and 20 μM) were assessed by CCK-8. (b). The H9C2 cell viability in control, H/R, and H/R + DEX were assessed by CCK-8. (c). FITC-PI kit was employed to assess cell apoptosis in H9C2 induced utilizing H/R or DEX. (d). Western blotting was utilized to measure Wnt3a, Wnt5a and β-catenin protein levels in H9C2 induced by H/R or DEX. CON, control; H/R, hypoxia/reoxygenation; Dex, dexmedetomidine. N = 3 for each group. *P < 0.05, **P < 0.001 vs. CON; #P < 0.05, ##P < 0.001 vs. H/R.
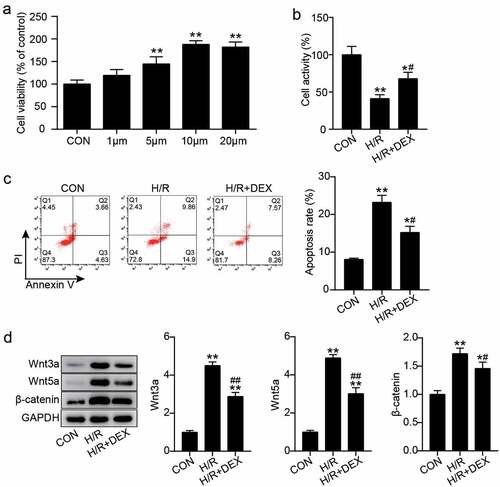
Figure 2. Knockdown of CCAT1 worsens H9C2 cell damage under H/R (a). CCAT1 level of H9C2 cells among control (CON), H/R, or DEX treated were assayed by qRT-PCR. **P < 0.001 vs. CON; ##P < 0.001 vs. H/R. (b). The expression levels of CCAT1 were detected by qRT-PCR in H9C2 cells treated with si-NC or si-CCAT1. (c). The cell viability of H9C2 cells in H/R, H/R + DEX, H/R + DEX +si-NC, and H/R + DEX +si-CCAT1 groups was assessed by CCK-8. (d). Flow cytometry was employed to assess cell apoptosis of H9C2 cells in H/R, H/R + DEX, H/R + DEX +si-NC, and H/R + DEX + si-CCAT1. (e). Western blotting was utilized to measure Wnt3a, Wnt5a and β-catenin protein levels in H9C2 treated with H/R, H/R + DEX, H/R + DEX +si-NC, and H/R + DEX + si-CCAT1. H/R, hypoxia/reoxygenation; Dex, dexmedetomidine; si-CCAT1, CCAT1 siRNA; si-NC, negative control of si-CCAT1. N = 3 for each group. **P < 0.001 vs. H/R; #P < 0.05, ##P < 0.001 vs. H/R + DEX +si-NC.
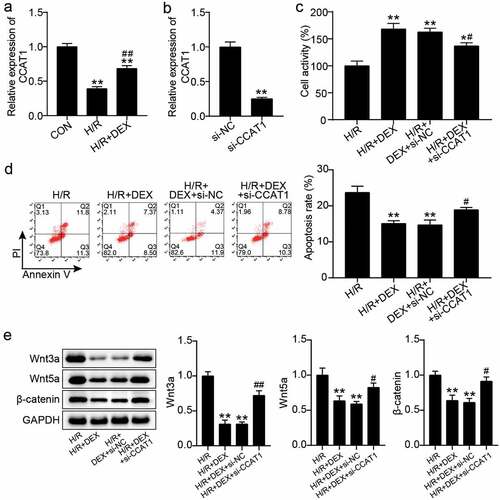
Figure 3. Up-regulation of CCAT1 promotes the reduction of H/R H9C2 cell injury induced by Dex treatment (a). The expression levels of CCAT1 were detected by qRT-PCR in H9C2 cells treated with empty vector or OE-CCAT1. (b). The cell viability of H9C2 cells in H/R, H/R + DEX, H/R + DEX + empty vector, and H/R + DEX + OE-CCAT1 groups was assessed by CCK-8. (c). Flow cytometry was employed to assess cell apoptosis of H9C2 cells in H/R, H/R + DEX, H/R + DEX + empty vector, and H/R + DEX + OE-CCAT1. (d). Western blotting was utilized to measure Wnt3a, Wnt5a and β-catenin protein levels in H9C2 treated with H/R, H/R + DEX, H/R + DEX + empty vector, and H/R + DEX + OE-CCAT1. CON, control; H/R, hypoxia/reoxygenation; Dex, dexmedetomidine; si-CCAT1, CCAT1 siRNA; si-NC, negative control of si-CCAT1. N = 3 for each group. **P < 0.001 vs. H/R; #P < 0.05, ##P < 0.001 vs. H/R + DEX + empty vector.
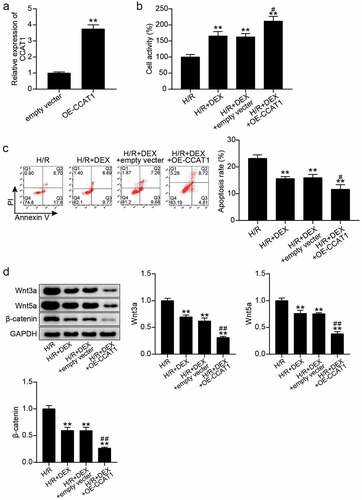
Figure 4. CCAT1 sponges miR-8063 (a). The predicted target sequences in CCAT1 which can directly bind with miR-8063. (b). Luciferase reporter assay was performed to indicate that miR-8063 could directly bind with CCAT1. **P < 0.001 vs. CCAT1-WT+miR-8063 mimic. (c). RIP assay was utilized to indicate the targeting relationship between CCAT1 and miR-8063. **P < 0.001 vs. IgG. (d). miR-8063 level of H9C2 cells among control (CON), H/R, or DEX treated were assayed by qRT-PCR. H/R, hypoxia/reoxygenation; Dex, dexmedetomidine. N = 3 for each group. **P < 0.001 vs. CON; ##P < 0.001 vs. H/R.
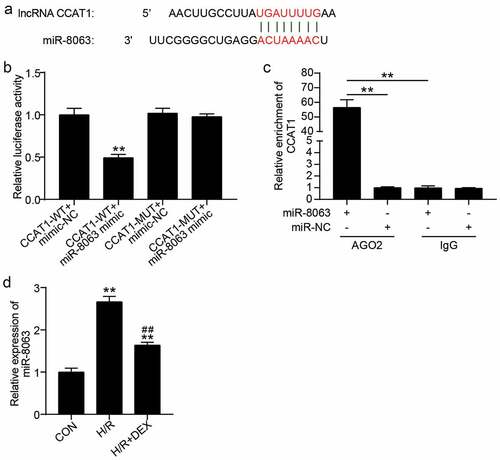
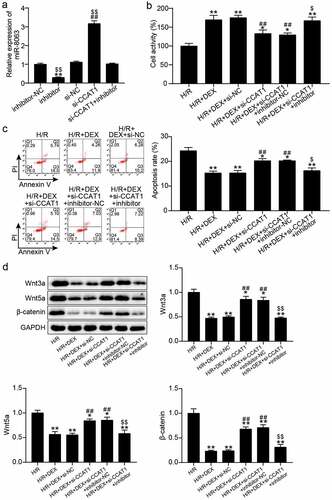
Figure 5. Interfering with miR-8063 reverse the effect of knockdown CCAT1 on the viability and apoptosis of H/R H9C2 cells (a). miR-8063 level of H9C2 cells among inhibitor-NC, inhibitor, si-NC, si-CCAT1, si-CCAT1 + inhibitor groups were assayed by qRT-PCR. (b). The cell viability of H9C2 cells in H/R, H/R + DEX, H/R + DEX +si-NC, H/R + DEX +si-CCAT1, H/R + DEX +si-CCAT1 + inhibitor-NC, and H/R + DEX +si-CCAT1 + inhibitor groups was assessed by CCK-8. (c). FITC-PI kit was employed to assess cell apoptosis in H/R, H/R + DEX, H/R + DEX +si-NC, H/R + DEX +si-CCAT1, H/R + DEX +si-CCAT1 + inhibitor-NC, and H/R + DEX +si-CCAT1 + inhibitor groups. (d). Western blotting was utilized to measure Wnt3a, Wnt5a and β-catenin protein levels in H9C2 treated with H/R, H/R + DEX, H/R + DEX +si-NC, H/R + DEX +si-CCAT1, H/R + DEX +si-CCAT1 + inhibitor-NC, and H/R + DEX +si-CCAT1 + inhibitor. H/R, hypoxia/reoxygenation; Dex, dexmedetomidine; si-CCAT1, CCAT1 siRNA; si-NC, negative control of si-CCAT1; inhibitor, miR-8063 inhibitor; inhibitor-NC, negative control of miR-8063 inhibitor. N = 3 for each group.