Figures & data
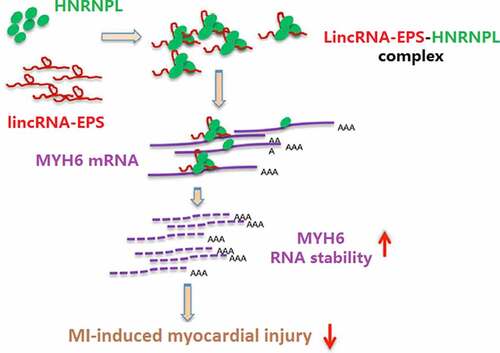
Figure 1. LincRNA-EPS relieves myocardial injury of MI mice. (a) The expressions of lincRNA-EPS in heart tissues were analyzed by qRT-PCR (n = 5). (b-g) The MI mice (n = 5) were intramyocardially injected with control vectors or lincRNA-EPS adenoviral vectors. (b) The expression of lincRNA-EPS was measured by qRT-PCR in heart tissues (n = 5). (c-g) The LVEF, LVFS, LVIDd and LVIDs were analyzed by Echocardiography analysis (n = 5). Data are presented as mean ± SD. Statistic significant differences were indicated: ** P < 0.01, # P < 0.05, ## P < 0.01.
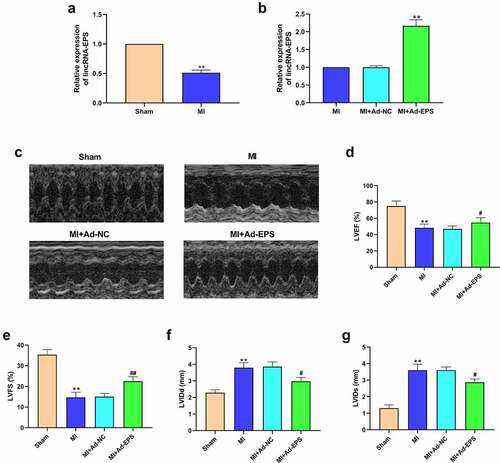
Figure 2. LincRNA-EPS attenuates inflammation and apoptosis in MI mice. (a-h) The MI mice (n = 5) were intramyocardially injected with control vectors or lincRNA-EPS adenoviral vectors. (a-d) The mRNA expressions of IL-6, TNF-α, IL-1β, and IL-18 were measured by qRT-PCR in mice. (e-h) The expressions of Bax, Bcl-2, caspase-3 and cleaved caspase-3 in heart tissues were assessed with Western blot. Data are presented as mean ± SD. Statistic significant differences were indicated: ** P < 0.01, ## P < 0.01.
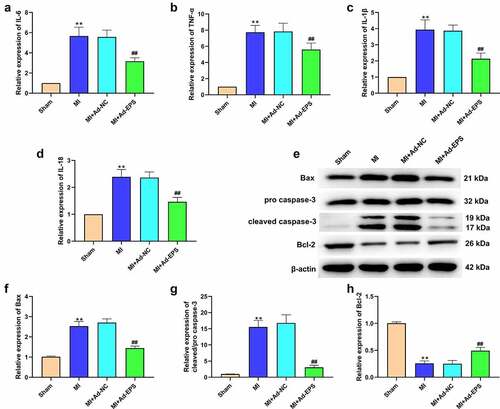
Figure 3. LincRNA-EPS inhibits OGD-induced cardiomyocyte injury in vitro. (a and b) HL-1 cells were treated with OGD for indicated times. (a) The viability of HL-1 cells was analyzed using MTT assay. (b) The level of lincRNA-EPS in HL-1 cells was tested using qRT-PCR. (c and d) The OGD-treated HL-1 cells and untreated HL-1 cells were transfected with pcDNA3.1 or pcDNA3.1 lincRNA-EPS overexpression vectors. (c) lincRNA-EPS level in HL-1 cells was tested using qRT-PCR. (d) LDH levels in culture medium of HL-1 cells were measured using colorimetric assay. Data are presented as mean ± SD. Statistic significant differences were indicated: * P < 0.05, ** P < 0.01, ## P < 0.01.
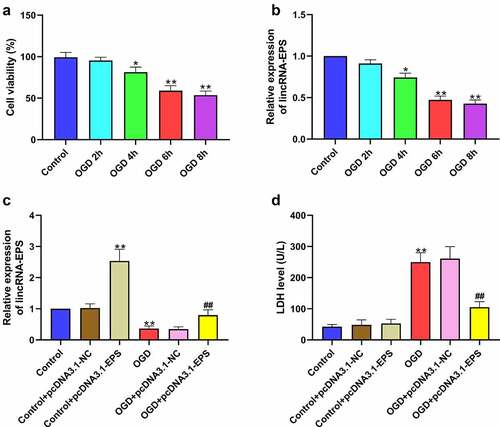
Figure 4. LincRNA-EPS alleviates OGD-induced inflammation and apoptosis in HL-1 cells. (a-i) The OGD-treated HL-1 cells and untreated HL-1 cells were treated with pcDNA3.1 or pcDNA3.1 lincRNA-EPS overexpression vectors. (a) The apoptosis of HL-1 cells was measured by TUNEL staining. (b-e) The protein expressions of Bax, Bcl-2, caspase-3 and cleaved caspase-3 in HL-1 cells were measured with Western blot. (f-i) The mRNA expressions of IL-6, TNF-α, IL-1β, and IL-18 in HL-1 cells were analyzed using qRT-PCR. Data are presented as mean ± SD. Statistic significant differences were indicated: ** P < 0.01, ## P < 0.01.
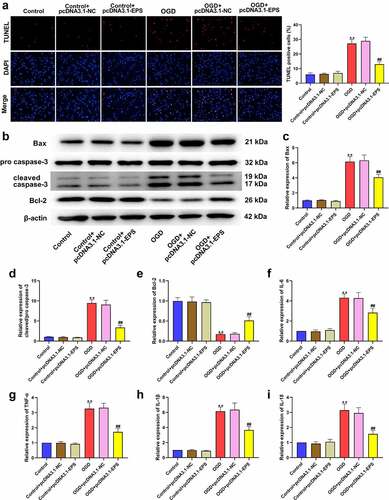
Figure 5. LincRNA-EPS enhances stability of MYH6 by interacting with HNRNPL in HL-1 cells. (a) The interaction of lincRNA-EPS, MYH6, and HNRNPL was analyzed by bioinformatics analysis. (b) The mRNA expression of HNRNPL was measured by qRT-PCR in the OGD-treated HL-1 cells. (c) The mRNA expression of MYH6 was tested by qRT-PCR in the OGD-treated HL-1 cells. (d) The interaction of lincRNA-EPS with HNRNPL and MYH6 was evaluated with RIP assay in HL-1 cells. (e and f) The interaction of HNRNPL with lincRNA-EPS and MYH6 was determined by RNA pull down assays in the HL-1 cells. (g) The HL-1 cells were treated with control shRNA, lincRNA-EPS shRNA or HNRNPL shRNA. The expression of lincRNA-EPS and HNRNPL was analyzed by qRT-PCR. (h) The HL-1 cells were co-treated with ActD and control shRNA, lincRNA-EPS shRNA, or HNRNPL shRNA. The mRNA expression of MYH6 was examined by qRT-PCR assays. (i) The HL-1 cells were treated with control shRNA or lincRNA-EPS shRNA. The expression of MYH6 was measured by Western blot. (j) The HL-1 cells were treated with control shRNA or HNRNPL shRNA. The expression of MYH6 was measured by Western blot. (k) The HL-1 cells were treated with pcDNA3.1 or pcDNA3.1 lincRNA-EPS overexpression vectors, or co-treated with pcDNA3.1 lincRNA-EPS overexpression vectors and HNRNPL shRNA. The expression of MYH6 was measured by Western blot. (l) The HL-1 cells were treated with pcDNA3.1 or pcDNA3.1 HNRNPL overexpression vectors. The expression of HNRNPL was measured by Western blot. (m) The HL-1 cells were co-treated with ActD and control shRNA, lincRNA-EPS shRNA, or co-transfected with lincRNA-EPS shRNA and pcDNA3.1 HNRNPL overexpression vectors. The mRNA expression of MYH6 was examined by qRT-PCR assays. Data are presented as mean ± SD. Statistic significant differences were indicated: ** P < 0.01, ## P < 0.01.
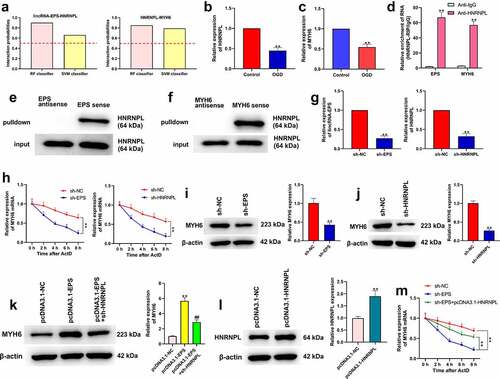
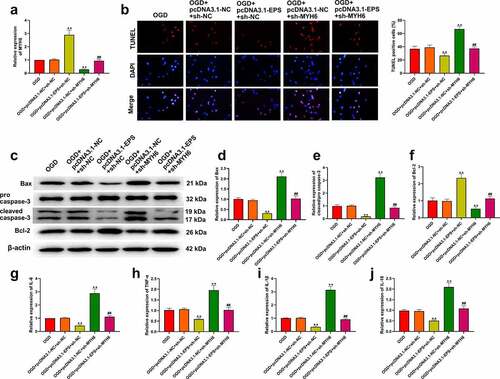
Figure 6. LincRNA-EPS represses OGD-induced inflammation and apoptosis by HNRNPL/MYH6 axis in HL-1 cells. (a-j) The OGD-treated HL-1 cells were treated with pcDNA3.1 or pcDNA3.1 lincRNA-EPS overexpression vectors, or co-treated with pcDNA3.1 lincRNA-EPS overexpression vectors and HNRNPL shRNA or MYH6 shRNA. (a) The expressions of MYH6 were tested using qRT-PCR. (b) The apoptosis of HL-1 cells was analyzed with TUNEL staining. (c-f) The expressions of Bax, Bcl-2, caspase-3 and cleaved caspase-3 were evaluated with Western blot. (g-j) The mRNA expression of IL-6, TNF-α, IL-1β, and IL-18 was tested using qRT-PCR. Data are presented as mean ± SD. Statistic significant differences were indicated: ** P < 0.01, ## P < 0.01.