Figures & data
Figure 1. The primary chondrocytes infected with Zfp521 were in good condition. (a) The effect of different doses of Zfp521 on chondrocyte viability was monitored in RTCA experiments. (b–c) Quantitative analysis of the cell index at 28 and 52 h. (d) The chondrocytes were observed using a fluorescence microscope in a pilot Zfp521 dose within an MOI range of 50–400. Scale bar: 100 μm. (e) Comparison of the rate of fluorescent positive cells between MOIs of 50 and 100. (f) The Zfp521 expression increased 8.304-fold at an MOI of 50 in the Ad-Zfp521 group compared with the vehicle group. (g) The Toluidine blue staining for the Ad-Zfp521 group at an MOI of 50 and the vehicle group. *P < 0.05, **P < 0.01, and ***P < 0.001. ns: not significant.
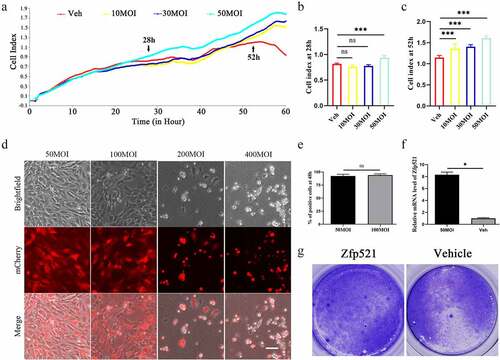
Figure 2. Here, Zfp521 affected the cell cycle progression and extracellular matrix–related pathways in the proteomics analysis (a) The hierarchical clustering of differential expression proteins. (b–c) Volcano plots and statistical plots of differential proteins. (d) Differential proteins were enriched in the pathway of the cell cycle phase and extracellular matrix in Biological Process of Gene Ontology analysis. (e) The cellular component analysis showed that differential proteins were enriched in the extracellular-associated region. (f) Network analysis for cyclin-dependent kinase-4 and MMP-13.
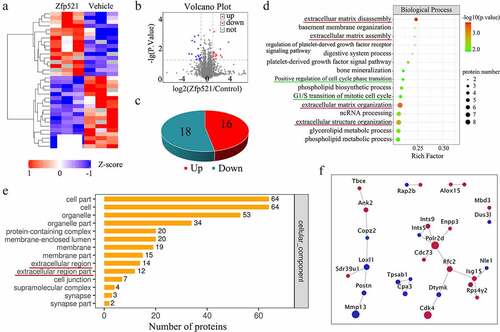
Figure 3. Here, Zfp521 promoted chondrocyte proliferation and inhibited apoptosis. (a) The EdU staining for the Zfp521-siRNA and vehicle siRNA chondrocytes was performed. Scale bar: 200 μm. (b) Comparison of the TUNEL staining for the Zfp521-siRNA and vehicle siRNA chondrocytes. Scale bar: 200 μm. (c-d) Quantitative analysis of the EdU and TUNEL assays between the Zfp521-siRNA group and vehicle siRNA group. (e–f) Immunohistochemical staining for PCNA and caspase-3 in the ACLT rats at nine weeks postop. Black scale bar: 50 μm. Blue scale bar: 15 μm. (g–h) Quantitative analysis of the immunohistochemical staining for PCNA and caspase-3 in the ACLT rats. **P < 0.01 and ***P < 0.001. ns: not significant.
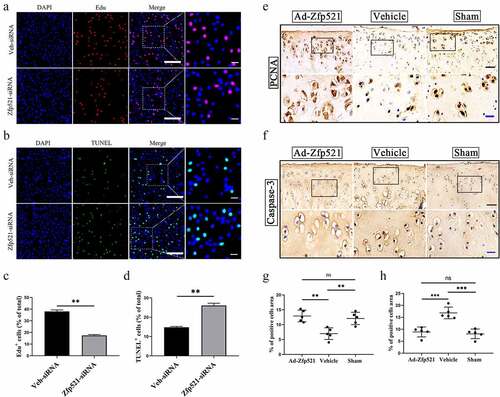
Figure 4. Here, Zfp521 suppressed the catabolic responses by downregulating Runx2 in the cartilage. (a) The RT-PCR of the IL1β-induced rat OA chondrocytes for catabolic biomarkers. (b) The total MMP levels in the living rats were monitored using FMT. (c) Quantitative comparison of FMT by the region of interest. (d) Immunohistochemical staining for Runx2, MMP-13, and Col10a1 at the protein level after nine weeks of treatment in the rats. Black scale bar: 50 μm. Blue scale bar: 15 μm. * P < 0.05. ns: not significant.
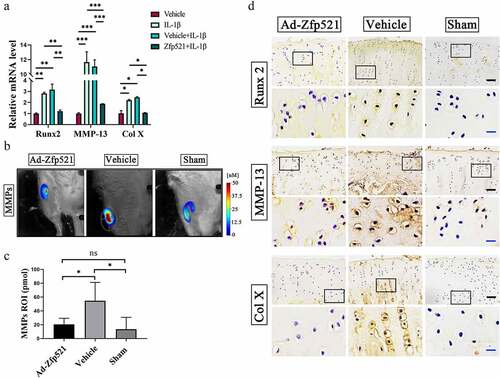
Figure 5. Here, Zfp521 enhanced anabolism by upregulating SOX9 in the cartilage. (a) The RT-PCR of the IL1β-induced rat OA chondrocytes for anabolic biomarkers. (b) Immunohistochemical staining for SOX9 and Col2a1 at the protein level after nine weeks of treatment in the rats. Black scale bar: 50 μm. Blue scale bar: 15 μm. (c–d) Safranin O/fast green staining and the OARSI scores for the cartilage evaluation. Scale bar: 50 μm. (e) Sample preparation for the nanoindentation test and labeling of the cartilage areas to be measured. (f–g) Young’s modulus of the cartilage in the medial and lateral tibial plateau. *P < 0.05.
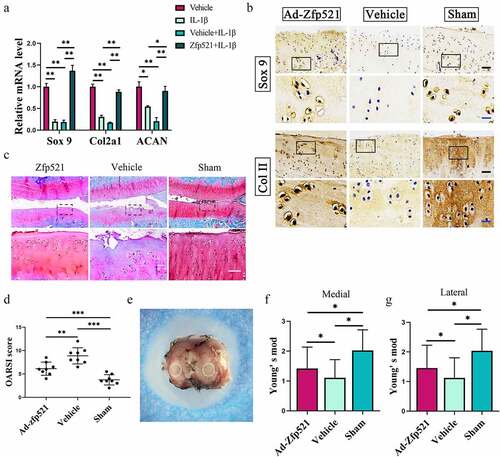
Figure 6. Here, Zfp521 relieved pain and attenuated knee degeneration. (a) Gait analysis for pain evaluation using the mean intensity/body weight of the right hind limb. (b–c) Quantitative comparison of the mean intensity/body weight and duty cycle. (d) India ink staining for the assessment of the surface erosion of the articular cartilage. (e) The severity of the osteoporosis and subchondral osteosclerosis were evaluated via X-rays. (f) Osteophytosis scores in the X-rays. * P < 0.05. ns: not significant.
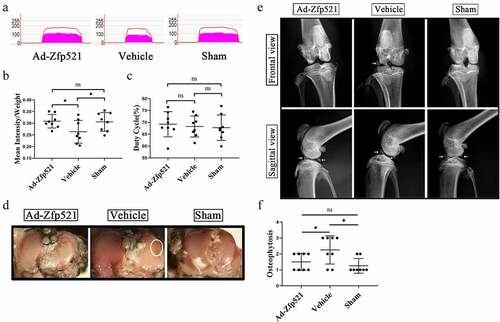
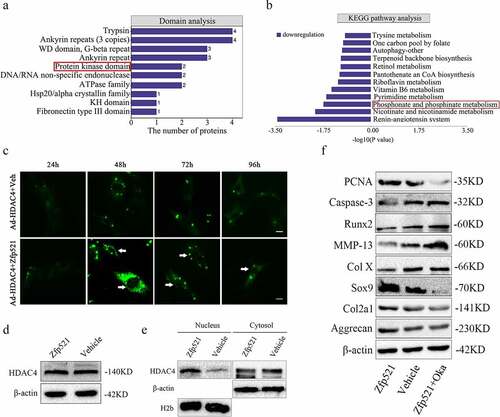
Figure 7. The molecular function of Zfp521 depended on the HDAC4 in the cell nucleus. (a) Domain analysis of proteomics for differentially expressed proteins. (b) A reduction in the phosphorylation metabolism in the KEGG pathway analysis. (c) The Zfp521 upregulation enabled HDAC4 to move into the nucleus at 48 h and onwards, as shown by the enhanced green fluorescence in the nucleus. Scale bar: 100 μm. (d) Here, Zfp521 did not affect the total protein expression of HDAC4. (e) The level of HDAC4 increased in the nucleus and decreased in the cytoplasm after Zfp521 upregulation. (f) The functions of Zfp521 were significantly weakened in the presence of okadaic acid.