Figures & data
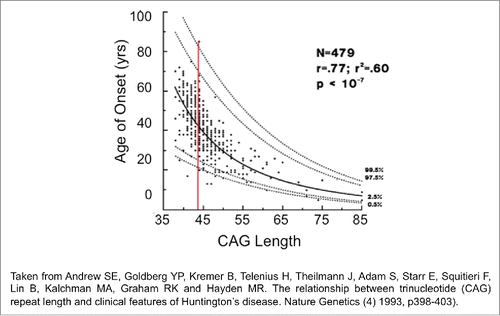
Figure 1. The mutation length distribution within tissues of Hdh(CAG)150 mice at 11 months. The histograms show a representative sample of CAG repeat lengths within the striatum, cortex, cerebellum and liver of an 11-month-old mouse. These data are compiled from small pool PCR analyses of reactions containing only 1–4 Alifiable mutant molecules in a few cells of one animal. The median CAG repeat length and number of mutant alleles (n) examined in each tissue is indicated. STR is striatum, CTX is cortex, CBL is cerebellum. Summing the change in tract length over many animals provides a large n at this particular age group.
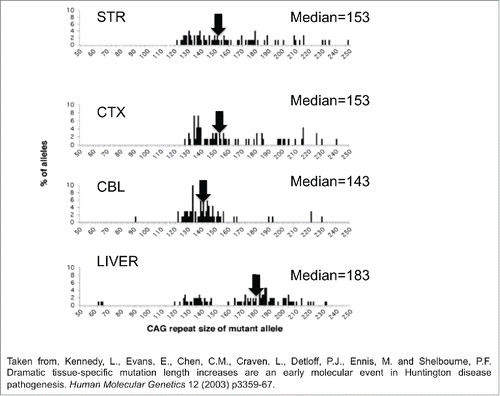
Figure 2. CAG repeat copy number of the progenitor mutation influences tissue-specific mutation length profiles. (A) Representative data from the SP–PCR mutation length profiles within striatum (St), cortex (Cx), cerebellum (Cb) and liver at 4 and 11 months of age in Hdh(Qwt/150) mice. Each lane contains the products from 1 to 10 cells worth of DNA from the tissue indicated. The numbers on the left hand side of the panels indicate the number of CAG repeats carried by the Hdh alleles amplified (mo=months). (B) Dramatic mutation length increases in human HD striata prior to pathological cell loss. Tissue from subject 1 shows mutation length variability in the striatum but not cortex (Brodmann's area 7) or hypothalamus. SP–PCR data reveal that while the median mutation length within tissues is ˜41 CAG repeats, some cells within the striatum contain mutations >1000 CAG repeats in length. Each lane contains the amplification products from ˜50 cells. (C,D) Tissue from subject 2 has mutation length variability in the striatum and cortex (Brodmann's area 7) but not cerebellum. SP–PCR data indicate the median mutation length within both tissues is ˜51 CAG repeats but some striatal cells contain mutations >700 CAG repeats in length. Each lane contains the amplification products from 40 to 50 cells. The numbers on the left side of each panel indicate the number of CAG repeats carried by the HD alleles amplified. The lanes marked M contain the 1Kb plus size marker.
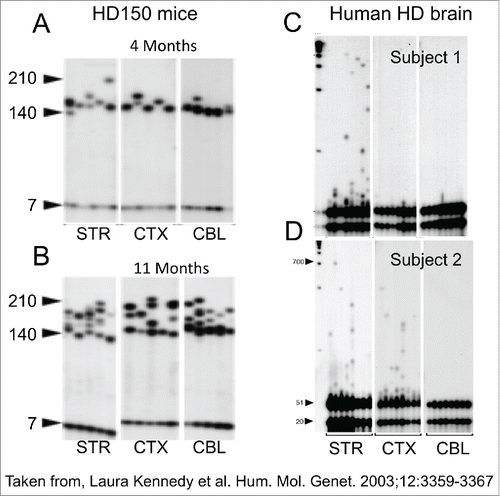
Figure 3. The wide distribution of behavioral responses for the same CAG repeat length in HD patients. The regression curve was calculated on log transformed data. Confidence intervals for predicted age given the size of the CAG repeat. Outer curves delineate the 99% confidence interval while inner curves show the 95% confidence interval.