Figures & data
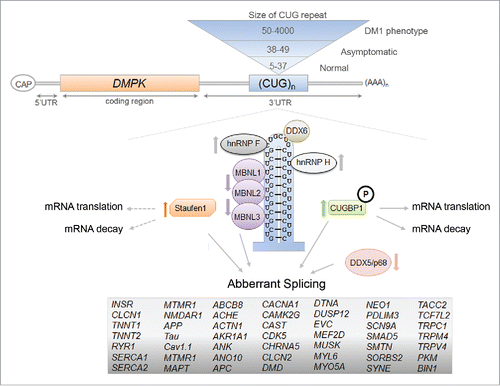
Figure 1. Diagram of the mammalian Staufen1 isoforms. All Stau1 isoforms contain the double-stranded RNA-binding domains (dsRBDs) 2, 3, 4, and 5 (orange boxes), the nuclear localization signal (NLS), the tubulin binding domain (TBD), and the reported Staufen-swapping motif (SSM) Citation65 (red diamond, dark gray and red boxes, respectively). The observed molecular weights are indicated in superscript and the amino acid positions are indicated in numbers.
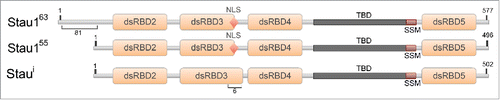
Figure 2. The proposed RNA-secondary structure of the IRAlus located in the INSR intron 10. The genomic DNA sequence of the human INSR (NG_008852.1) was used to assess the Alu repeat elements located in intron 10. Alu elements were identified using RepeatMasker and RNA secondary structure was determined by Vienna package RNAfold 2.1.1. The intronic regions shown here are not to scale and this is indicated by a // symbol.
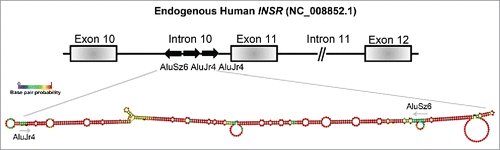
Figure 3. The RNA-binding proteins that are involved in the molecular pathomechanisms of Myotonic Dystrophy Type 1. The toxic RNA-gain of function model of DM1 that shows the expansion of CUG-repeat mRNA in the nucleus and the resulting misregulation of RBPs. The RBPs that can act as disease modifiers in the DM1 pathology, through the regulation of alternative splicing events, mRNA translation and decay, are shown here and the arrows represent the decrease/increase in either protein levels and/or activities of the protein in DM1 (references within main text). The mRNAs containing aberrant splicing events that have been identified in various DM1 models are listed.