Figures & data
Table 1. Overview of different phenotypes observed after Jacob/Nsmf knockdown or knockout approaches with respect to KS.
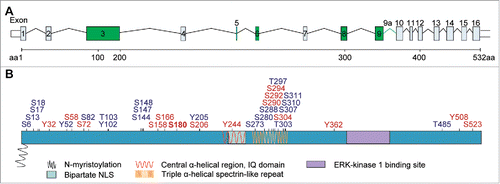
Figure 1. Translocation of Jacob/Nsmf to the nucleus is a key factor for a positive feedback loop involved in BDNF synthesis. BDNF induces the NMDAR-dependent translocation of phosphorylated Jacob to the nucleus in a trimeric complex with pERK1/2 and α-internexin. Higher levels of nuclear pJacob and pERK1/2 substantially contribute to expression of CREB-dependent genes including bdnf. BDNF synthesis enhances dendritic and synaptic development, necessary for unaltered synapto-nuclear communication, cell survival and expression of plasticity-related genes.
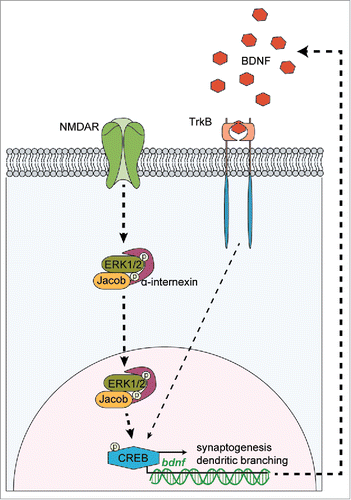
Figure 2. Genomic structure of the mouse Jacob/Nsmf gene, key motifs and phosphorylation sites of the Jacob protein. (A) The Nsmf mouse gene consists of 16 exons. Exons 3, 5, 6, 8, 9 (marked in green) can be alternatively spliced. In addition, intron 9 has been predicted to constitute for one further isoform (denoted 9a) (B). The Jacob/Nsmf protein is largely unstructured but contains several motifs like a N-myristoylation site, a bipartite NLS, an IQ domain, central α-helical region, ERK-1 kinase binding site, and Triple α-helical spectrin-like repeats described before. Disorder Enhanced Phosphorylation Predictor (DEEP) revealed numerous phosphorylation sites (blue; only phosphorylation sites with DEEP score above 0.7 were included). Analysis of Jacob mouse protein with PhosphoSitePlus tool (Cell Signaling) revealed numerous phosphorylation sites reported by more than one Mass Spectrometry analysis studies (red). In bold, a S180 phosphorylation site confirmed by site-specific method, i.e. site-directed mutagenesis, mass-spectrometry and specific antibodies.Citation25-27