Figures & data
Figure 1 A novel PCR technology for the amplification of the hexanucleotide G4C2 repeat element in the C9orf72 gene. (a) Schematic representation of the C9orf72 gene structure showing the predicted 11 exons (boxes), the location of the intronic hexanucleotide repeat expansion (vertical lines) and the 3′-SV region (black triangle). (b) Schematics of the 3-primer GS/RP-PCR design (dot—FAM fluorophore) and (c) 2-primer GS-PCR design. (d) A representative 3-primer GS/RP-PCR/CE profile for an expanded Coriell sample (ND10206) that overlays GS peaks (a normal allele at five repeats, an expanded allele at 54 repeats) with a hyper-expanded RP-PCR profile and corresponding pile-up peak (inset, >145 repeats). (e) Matching 2-primer GS-PCR products for sample ND10206 resolved on CE and AGE (inset) shows the two alleles (sized at five and 54 repeats). The hyper-expanded pile-up peak is resolved in the gel image as a 502 and a 597 repeat.
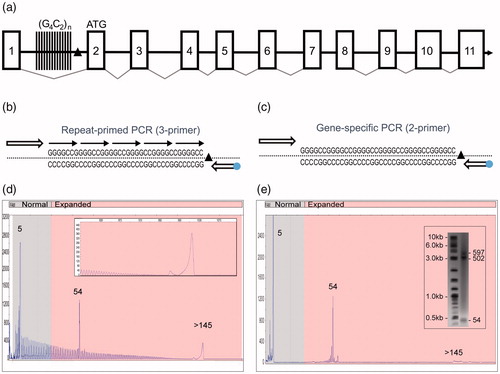
Figure 2 Performance characteristics of the C9orf72 PCR/CE assay. (a) Accurate sizing of up to 145 repeats with a distinct RP-PCR profile for expanded samples is achieved using the 3-primer PCR design. The RP profile enables zygosity resolution, i.e. distinguishing a homozygous sample with two alleles with seven repeats (ND09188; left) and an expanded heterozygous sample (ND12102; right) with one allele with seven repeats and a hyper-expanded allele. (b) The 3-primer PCR demonstrates robust performance and detection of expanded alleles across a 200-fold gDNA range down to 1 ng/reaction (ND12754). Long-range RP peak counting is denoted at the 100, 120, and 140 repeat peaks. (c) The presence of expanded allele RP profiles are visible down to a 5% mass fraction (admixture of expanded ND10966 in the background of unexpanded ND09188). (d) Low-level minor alleles are readily detected in the 2-primer assay configuration on both CE and AGE (ND10689; left, inset) as compared to the 3-primer assay (right). Numbers represent allele sizes (repeats—RPs).
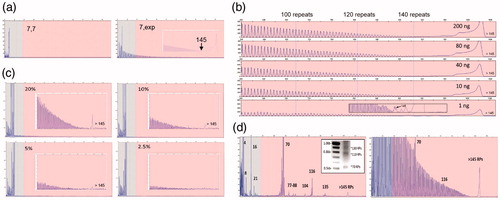
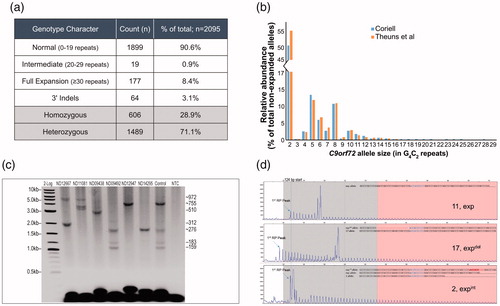
Figure 3 Comprehensive C9orf72 genotyping of 2095 NINDS ALS samples from the Coriell cell repository. (a) Genotype summary table. (b) The normal and intermediate C9orf72 repeat size distribution (by allele; <30 repeats) within the tested Coriell NINDS ALS sample set was consistent with previous reports (Citation12). (c) Representative GS-PCR results, repeat sizes are listed (right). Detection of hyper-expanded ∼5.8 kb (950 repeats, ND12667, ND11081) amplicons and low-level size mosaicism. An admixture of samples ND06751 and ND09492 in equal parts was used to assess processivity, sensitivity, and AGE sizing accuracy. (d) A noticeable offset in the RP single start site (∼124 bp), or signal dips in the RP profile are evident when 3′ deletions or insertions are present. Representative samples and their confirmed underlying sequence are shown. Top pane (no 3′-SV), ND09438; middle pane (deletion), ND12947; bottom pane (insertion), ND09492.