Figures & data
Table 1 Commercial clinical genetic tests offered globally specific to amyotrophic lateral sclerosis (ALS) and motor neuron disease.
Figure 1 Year of first discovery of a relationship with amyotrophic lateral sclerosis (ALS), method of discovery, and function of the encoded protein of the genes included on the identified ALS specific commercial clinical genetic tests (N = 14) with ≥2 publications reporting rare variants associated with ALS.
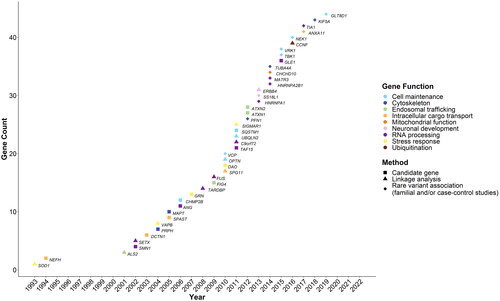
Figure 2 Packed circle plot comparing the number of amyotrophic lateral sclerosis (ALS) specific commercial clinical genetic tests (N = 14) each gene was included on. The size of the circles corresponds to the number of clinical genetic tests the gene was included on, ranging from one to 14 tests. The color of the circles corresponds to the number of years since the gene’s first association with ALS was made. Grey circles indicate genes that have never been associated with ALS. The year of first association between ALS and the gene is indicated below the gene name, where applicable.
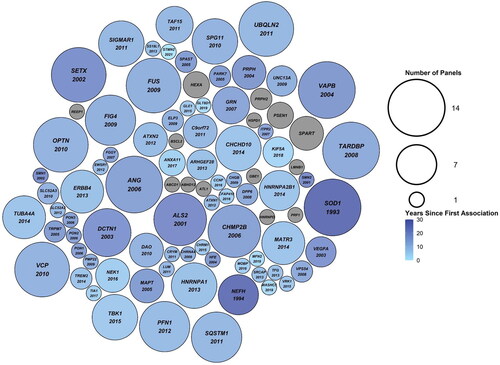
Table 2 Genes covered by the commercial clinical genetic tests offered globally specific to amyotrophic lateral sclerosis (ALS).
Table 3 Characteristics of the genes encompassed by the commercial clinical genetic tests offered globally specific to amyotrophic lateral sclerosis (ALS), as well as potential genes of interest missing from clinical genetic tests.
Figure 3 Pathogenicity classification and variant types reported in ClinVar in genes previously associated with amyotrophic lateral sclerosis (ALS). All genes found on at least one of the analyzed clinical genetic testing panels (N = 14) were included in the analysis. However, only genes with variants reported as associated with “ALS” or “motor neuron disease” in ClinVar are displayed in the figure. (A) Flow chart highlighting the steps taken to obtain and filter the ClinVar variants reported in genes previously associated with ALS. (B) Pathogenicity classification of all variants reported in ClinVar as associated with ALS. The total number of variants reported in ClinVar in DCTN1, FUS, MATR3, SETX, SQSTM1, and VAPB exceeded the y-axis limits of 200; for these genes, the total number of variants found in ClinVar were included on the right side of the bar plot. (C) Variant types of all variants reported in ClinVar as likely pathogenic or pathogenic for ALS. (D) Variant types of all variants reported in ClinVar as being of uncertain significance for ALS. Abbreviations: LP, likely pathogenic; P, pathogenic; VUS, variants of uncertain significance.

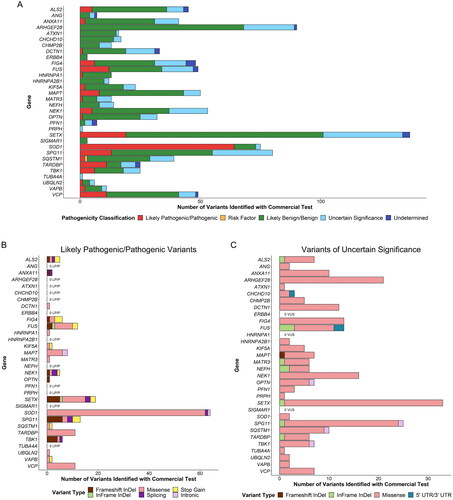
Figure 4 Pathogenicity classification and variant types identified using a commercially available genetic test for amyotrophic lateral sclerosis (ALS). (A) Pathogenicity classification of variants identified using a commercially available genetic test for ALS. (B) Variant types of all variants identified using a commercially available genetic test for ALS and classified as likely pathogenic or pathogenic. (C) Variant types of all variants identified using a commercially available genetic test for ALS and classified as being of uncertain significance. Abbreviations: LP, likely pathogenic; P, pathogenic; VUS, variants of uncertain significance.