Figures & data
Figure 1. Representative left eye images of 32 year old male with CHM mutation. (a) Widefield optos color image showing baring of sclera and peripheral pigmentation. (b) Macular fundus autofluorescence showing the central area of relative RPE preservation. (c) OCT image showing loss of outer retina with central island of preservation. (d) Microperimetry data showing central preservation of retinal sensitivity with sharply demarcated sensitivity drop-off corresponding to island of surviving RPE
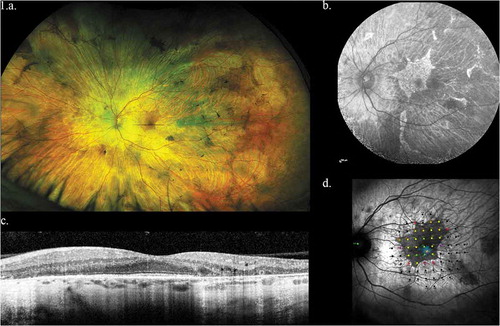
Figure 2a. (a) Structure of the CHM gene annotated with pathogenic variants. Intronic mutations have been color coded with a pink header, exonic mutations with a blue header. Single-base substitutions are shown in bold: of these, single-base substitutions resulting in missense mutations are colored red; terminations are colored green, and splicing defects are colored purple. The five most common described variants are underlined (c.(?-30)_(*3450?)del (n = 20), c.757 C > T (n = 18), c.799 C > T (n = 16), c.1584_1587del (n = 11), 808 C > T and 877 C > T (n = 10 each)
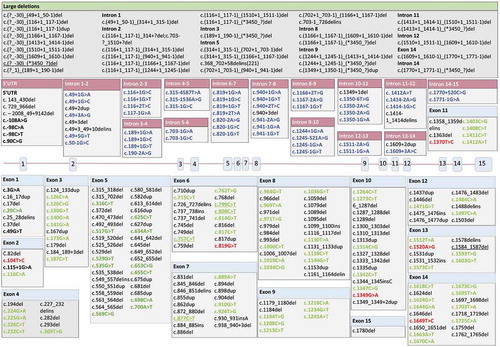
Figure 2b. (b) Classification of pathogenic variants: inner ring shows mutation location (exonic, intronic, multiple locations (comprising large or multiple indels or duplications); outer ring shows effect of mutation (premature termination codon, missense variant, no protein product formed, frameshift, unknown) Source: Leiden Open Variation Database (LOVD) [Citation27]
![Figure 2b. (b) Classification of pathogenic variants: inner ring shows mutation location (exonic, intronic, multiple locations (comprising large or multiple indels or duplications); outer ring shows effect of mutation (premature termination codon, missense variant, no protein product formed, frameshift, unknown) Source: Leiden Open Variation Database (LOVD) [Citation27]](/cms/asset/9f9457f3-67f8-422b-8e2e-0b5e42a5f46a/ieod_a_1882300_f0002b_oc.jpg)
Table 1. Summary of trials involving REP1 replacement and their status
Figure 3. Potential genome editing targets in CHM. The inner ring shows type of mutation. The middle ring shows the single-base substitutions (which overwhelmingly produce terminations, as previously described) and other consequences of the mutations (Frameshifts, splice defects, no protein product formed, or unknown). The outer ring shows the number of potential base editing targets in CHM (red: currently restricted to transitions, which comprise approximately 30.8% of choroideremia-associated mutations; the remaining 69.2% are not amenable to base-editing methods): the number of potential prime editing (orange: prime editing opens up the potential number of targets up to 85.7% of CHM variants): and remaining uneditable mutations (yellow: large insertions, deletions and duplications) Source: Leiden Open Variation Database (LOVD) [Citation27]
![Figure 3. Potential genome editing targets in CHM. The inner ring shows type of mutation. The middle ring shows the single-base substitutions (which overwhelmingly produce terminations, as previously described) and other consequences of the mutations (Frameshifts, splice defects, no protein product formed, or unknown). The outer ring shows the number of potential base editing targets in CHM (red: currently restricted to transitions, which comprise approximately 30.8% of choroideremia-associated mutations; the remaining 69.2% are not amenable to base-editing methods): the number of potential prime editing (orange: prime editing opens up the potential number of targets up to 85.7% of CHM variants): and remaining uneditable mutations (yellow: large insertions, deletions and duplications) Source: Leiden Open Variation Database (LOVD) [Citation27]](/cms/asset/0b0cbc3b-996c-45cf-b57f-0ad59021b219/ieod_a_1882300_f0003_oc.jpg)
Table 2. Most common substitutions in CHM. The top five are all C to T transitions, which lead to the creation of a STOP codon
