Figures & data
Figure 1. Detection of tight junction proteins in oral (buccal, tonsil), cervical and intestinal (jejunal region) epithelia. Representative immunofluorescence images of tight junction proteins occludin (red) in multistratified buccal, tonsil, and ectocervical mucosal epithelia and ZO-1 (red) in a single-cell layer of intestinal epithelium are shown. Oral (buccal and tonsil) squamous mucosal epithelia have well-developed tight junctions in the strata granulosum, spinosum, and parabasal layers. Cervical epithelia form tight junctions in the strata spinosum and parabasal layers. A single-cell layer of intestinal epithelium also has well-developed tight junctions. The tight junctions are localized at the upper lateral membranes of epithelial cells and seal intercellular spaces, preventing paracellular penetration by viruses. Nuclei are stained blue. Dashed white lines separate epithelium from the lamina propria. GR, granulosum; SP, spinosum; BL, basal; LP, lamina propria; EP, epithelium. Confocal microscopy. Original magnification: for buccal, tonsil and cervical x400, for intestinal x630
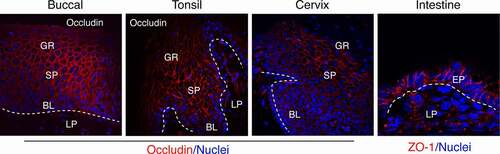
Figure 2. Model of organization of epithelial tight junctions and their disruption. Tight junctions are formed between epithelial cells of oral, genital, and intestinal mucosa by lateral interaction of integral membrane proteins occludin and claudins, which are associated with the cytoplasmic proteins ZO-1, −2, and −3 (left). These proteins link occludin and claudins to the actin cytoskeleton. Adherens junctions are formed in the lateral membranes of epithelial cells by homophilic interactions of E-cadherins. Junctional adhesion molecules (JAMs), coxsackievirus and adenovirus receptor (CAR), and nectin-1 are also localized at the tight and adherens junctions of mucosal epithelia. They may serve as receptors for reovirus, coxsackievirus/adenovirus, and HSV-1, respectively, and are sequestered within the tight and adherens junctions. Interactions of viral pathogens with mucosal epithelial cells may directly or indirectly cause disruption of epithelial junctions by downregulation of junctional protein expression and/or their mislocalization from assembled junctions, leading to dissociation of tight junction proteins from each other and from actin cytoskeleton (right). Disruption of epithelial tight junctions facilitates (i) opening of the paracellular space between epithelial cells and (ii) paracellular penetration by viruses. Disruption of tight and adherens junctions may release sequestered JAMs, CAR, and nectin-1, which may increase viral accessibility
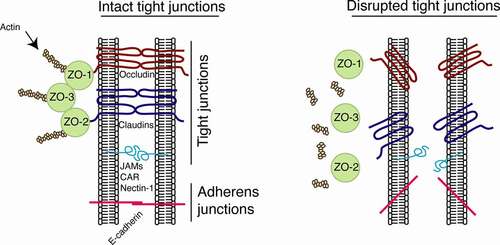
Table 1. Interaction of human pathogenic viruses with mucosal epithelial junctional proteins
